Rain Tomorrow
By Charles Holbert
August 28, 2022
For this post, we will evaluate rainfall in Australia using daily weather observations from multiple Australian weather stations. The data include a target variable RainTomorrow, which identifies (Yes/No) whether there was rain on the following day. We will build several machine learning models using the tidymodels framework for use in predicting if there will be rain tomorrow. Models will include logistic regression, random forest, extreme gradient boosted trees, and a neural network.
Load Libraries
Load libraries and any functions that may be required to analyze the data.
# Load libraries
library(dplyr)
library(tidyr)
library(ggplot2)
library(scales)
library(reshape2)
library(tidyquant)
library(ggcorrplot)
library(vip)
source('functions/outlier_functions.R')
Data
Read Data
Load the data from the csv file and inspect data structure. This dataset contains over 140,000 daily observations from over 45 Australian weather stations. RainTomorrow is the target variable to predict. If this column is Yes, rain for the next day was 1 mm or more.
# Read data in comma-delimited file
dat <- read.csv('data/weatherAUS.csv', header = T)
# Read location data in comma-delimited file
locs <- read.csv('data/locations.csv', header = T)
# Inspect structure of data
str(dat)
## 'data.frame': 145460 obs. of 23 variables:
## $ Date : chr "2008-12-01" "2008-12-02" "2008-12-03" "2008-12-04" ...
## $ Location : chr "Albury" "Albury" "Albury" "Albury" ...
## $ MinTemp : num 13.4 7.4 12.9 9.2 17.5 14.6 14.3 7.7 9.7 13.1 ...
## $ MaxTemp : num 22.9 25.1 25.7 28 32.3 29.7 25 26.7 31.9 30.1 ...
## $ Rainfall : num 0.6 0 0 0 1 0.2 0 0 0 1.4 ...
## $ Evaporation : num NA NA NA NA NA NA NA NA NA NA ...
## $ Sunshine : num NA NA NA NA NA NA NA NA NA NA ...
## $ WindGustDir : chr "W" "WNW" "WSW" "NE" ...
## $ WindGustSpeed: int 44 44 46 24 41 56 50 35 80 28 ...
## $ WindDir9am : chr "W" "NNW" "W" "SE" ...
## $ WindDir3pm : chr "WNW" "WSW" "WSW" "E" ...
## $ WindSpeed9am : int 20 4 19 11 7 19 20 6 7 15 ...
## $ WindSpeed3pm : int 24 22 26 9 20 24 24 17 28 11 ...
## $ Humidity9am : int 71 44 38 45 82 55 49 48 42 58 ...
## $ Humidity3pm : int 22 25 30 16 33 23 19 19 9 27 ...
## $ Pressure9am : num 1008 1011 1008 1018 1011 ...
## $ Pressure3pm : num 1007 1008 1009 1013 1006 ...
## $ Cloud9am : int 8 NA NA NA 7 NA 1 NA NA NA ...
## $ Cloud3pm : int NA NA 2 NA 8 NA NA NA NA NA ...
## $ Temp9am : num 16.9 17.2 21 18.1 17.8 20.6 18.1 16.3 18.3 20.1 ...
## $ Temp3pm : num 21.8 24.3 23.2 26.5 29.7 28.9 24.6 25.5 30.2 28.2 ...
## $ RainToday : chr "No" "No" "No" "No" ...
## $ RainTomorrow : chr "No" "No" "No" "No" ...
Prep Data
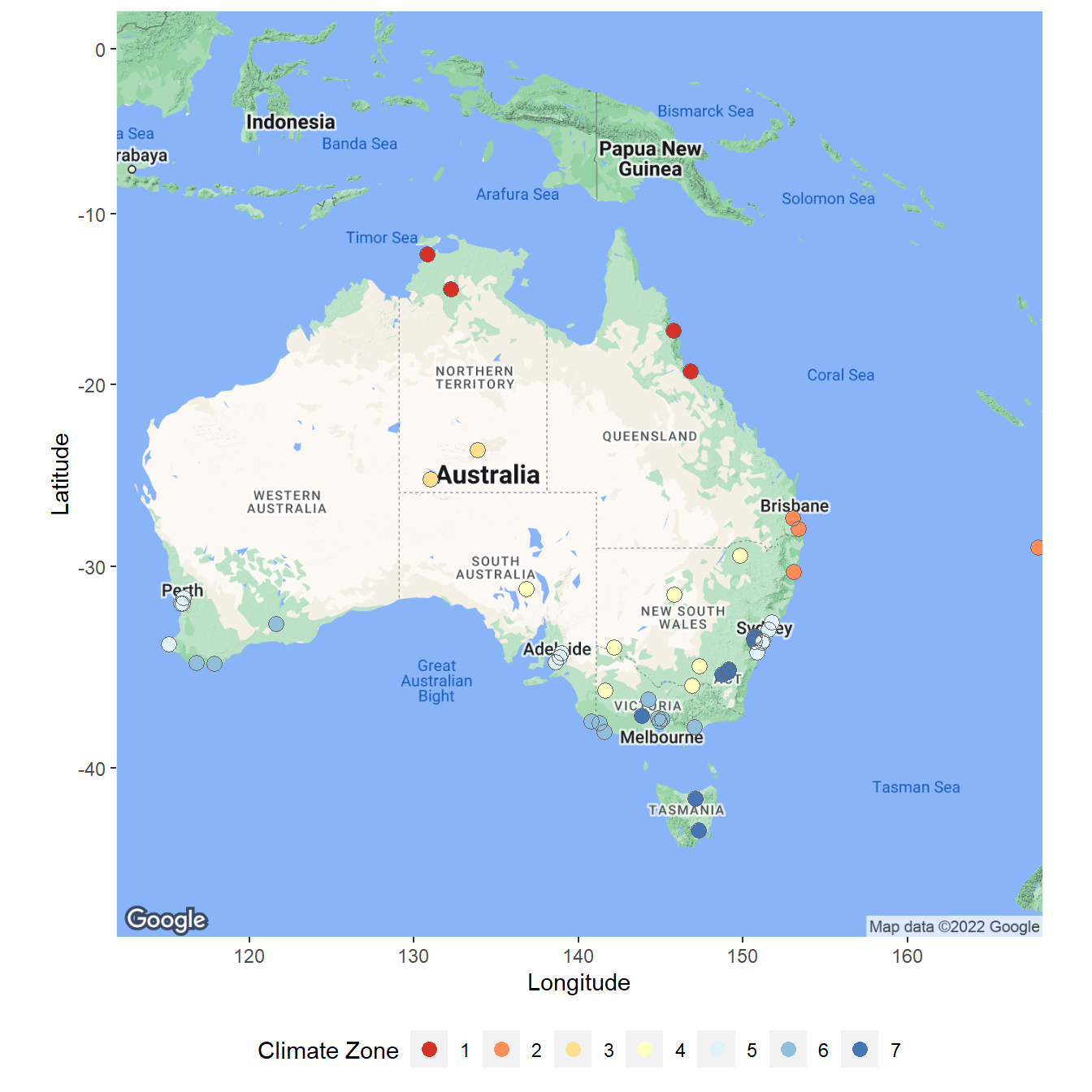
Let’s prep the data prior to modeling. First, we will join the data with a locations file that contains geographical coordinates and an assigned climate zone for each weather station. Locations with approximately similar climates have been combined into eight climate zones, which include the following:
- Climate zone 1 - high humidity summer, warm winter
- Climate zone 2 - warm humid summer, mild winter
- Climate zone 3 - hot dry summer, warm winter
- Climate zone 4 - hot dry summer, cool winter
- Climate zone 5 - warm temperate
- Climate zone 6 - mild temperate
- Climate zone 7 - cool temperate
- Climate zone 8 - alpine
Let’s also add some additional variables to the data, including month, year, and differential humidity, temperature, and pressure.
# Add variables
dat <- dat %>%
left_join(locs, by = 'Location') %>%
mutate(
Date = as.Date(dat$Date),
Month = lubridate::month(Date, label = TRUE),
Year = as.factor(lubridate::year(Date)),
diffHumidity = Humidity3pm - Humidity9am,
diffTemp = Temp3pm - Temp9am,
diffPressure = Pressure3pm - Pressure9am
)
Create Location Map
Let’s create a map showing the locations of the weather stations color coded by climate zone.
# Get map of Australia
myMap <- get_map(location = c(140, -25), zoom = 4, scale = 2)
# Create map
ggmap(myMap) +
geom_point(data = locs, aes(x = Lng, y = Lat,
color = as.factor(Zone)), shape = 16, size = 3) +
geom_point(data = locs, aes(x = Lng, y = Lat), color = 'grey40',
shape = 1, size = 3.3, stroke = 0.2) +
labs(x = 'Longitude',
y = 'Latitude') +
scale_color_brewer(palette = 'RdYlBu') +
guides(color = guide_legend(title = 'Climate Zone', nrow = 1)) +
theme(legend.position = 'bottom')
Inspect Data
Let’s inspect the overall structure of the data and get a summary for all the variables.
# Summary of data using skimr
skimr::skim(dat)
Table: Table 1: Data summary
| Name | dat |
| Number of rows | 145460 |
| Number of columns | 31 |
| _______________________ | |
| Column type frequency: | |
| character | 6 |
| Date | 1 |
| factor | 2 |
| numeric | 22 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| Location | 0 | 1.00 | 4 | 16 | 0 | 49 | 0 |
| WindGustDir | 10326 | 0.93 | 1 | 3 | 0 | 16 | 0 |
| WindDir9am | 10566 | 0.93 | 1 | 3 | 0 | 16 | 0 |
| WindDir3pm | 4228 | 0.97 | 1 | 3 | 0 | 16 | 0 |
| RainToday | 3261 | 0.98 | 2 | 3 | 0 | 2 | 0 |
| RainTomorrow | 3267 | 0.98 | 2 | 3 | 0 | 2 | 0 |
Variable type: Date
| skim_variable | n_missing | complete_rate | min | max | median | n_unique |
|---|---|---|---|---|---|---|
| Date | 0 | 1 | 2007-11-01 | 2017-06-25 | 2013-06-02 | 3436 |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| Month | 0 | 1 | TRUE | 12 | Mar: 13361, May: 13353, Jan: 13236, Jun: 12684 |
| Year | 0 | 1 | FALSE | 11 | 201: 17934, 201: 17885, 201: 17885, 200: 16789 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| MinTemp | 1485 | 0.99 | 12.19 | 6.40 | -8.50 | 7.60 | 12.00 | 16.90 | 33.90 | ▁▅▇▅▁ |
| MaxTemp | 1261 | 0.99 | 23.22 | 7.12 | -4.80 | 17.90 | 22.60 | 28.20 | 48.10 | ▁▂▇▅▁ |
| Rainfall | 3261 | 0.98 | 2.36 | 8.48 | 0.00 | 0.00 | 0.00 | 0.80 | 371.00 | ▇▁▁▁▁ |
| Evaporation | 62790 | 0.57 | 5.47 | 4.19 | 0.00 | 2.60 | 4.80 | 7.40 | 145.00 | ▇▁▁▁▁ |
| Sunshine | 69835 | 0.52 | 7.61 | 3.79 | 0.00 | 4.80 | 8.40 | 10.60 | 14.50 | ▃▃▅▇▃ |
| WindGustSpeed | 10263 | 0.93 | 40.04 | 13.61 | 6.00 | 31.00 | 39.00 | 48.00 | 135.00 | ▅▇▁▁▁ |
| WindSpeed9am | 1767 | 0.99 | 14.04 | 8.92 | 0.00 | 7.00 | 13.00 | 19.00 | 130.00 | ▇▁▁▁▁ |
| WindSpeed3pm | 3062 | 0.98 | 18.66 | 8.81 | 0.00 | 13.00 | 19.00 | 24.00 | 87.00 | ▇▇▁▁▁ |
| Humidity9am | 2654 | 0.98 | 68.88 | 19.03 | 0.00 | 57.00 | 70.00 | 83.00 | 100.00 | ▁▁▅▇▆ |
| Humidity3pm | 4507 | 0.97 | 51.54 | 20.80 | 0.00 | 37.00 | 52.00 | 66.00 | 100.00 | ▂▅▇▆▂ |
| Pressure9am | 15065 | 0.90 | 1017.65 | 7.11 | 980.50 | 1012.90 | 1017.60 | 1022.40 | 1041.00 | ▁▁▇▇▁ |
| Pressure3pm | 15028 | 0.90 | 1015.26 | 7.04 | 977.10 | 1010.40 | 1015.20 | 1020.00 | 1039.60 | ▁▁▇▇▁ |
| Cloud9am | 55888 | 0.62 | 4.45 | 2.89 | 0.00 | 1.00 | 5.00 | 7.00 | 9.00 | ▇▃▃▇▅ |
| Cloud3pm | 59358 | 0.59 | 4.51 | 2.72 | 0.00 | 2.00 | 5.00 | 7.00 | 9.00 | ▆▅▃▇▃ |
| Temp9am | 1767 | 0.99 | 16.99 | 6.49 | -7.20 | 12.30 | 16.70 | 21.60 | 40.20 | ▁▃▇▃▁ |
| Temp3pm | 3609 | 0.98 | 21.68 | 6.94 | -5.40 | 16.60 | 21.10 | 26.40 | 46.70 | ▁▃▇▃▁ |
| Lng | 0 | 1.00 | 141.83 | 12.02 | 115.10 | 138.60 | 145.77 | 150.69 | 167.95 | ▂▁▆▇▁ |
| Lat | 0 | 1.00 | -32.81 | 6.01 | -42.88 | -36.08 | -34.02 | -31.50 | -12.44 | ▃▇▂▁▁ |
| Zone | 0 | 1.00 | 4.86 | 1.79 | 1.00 | 4.00 | 5.00 | 6.00 | 7.00 | ▃▁▂▅▇ |
| diffHumidity | 5274 | 0.96 | -17.35 | 16.33 | -91.00 | -28.00 | -17.00 | -6.00 | 91.00 | ▁▇▇▁▁ |
| diffTemp | 4247 | 0.97 | 4.76 | 3.56 | -12.60 | 2.10 | 4.40 | 7.20 | 23.00 | ▁▂▇▂▁ |
| diffPressure | 15289 | 0.89 | -2.40 | 1.97 | -16.70 | -3.70 | -2.60 | -1.30 | 10.80 | ▁▁▇▁▁ |
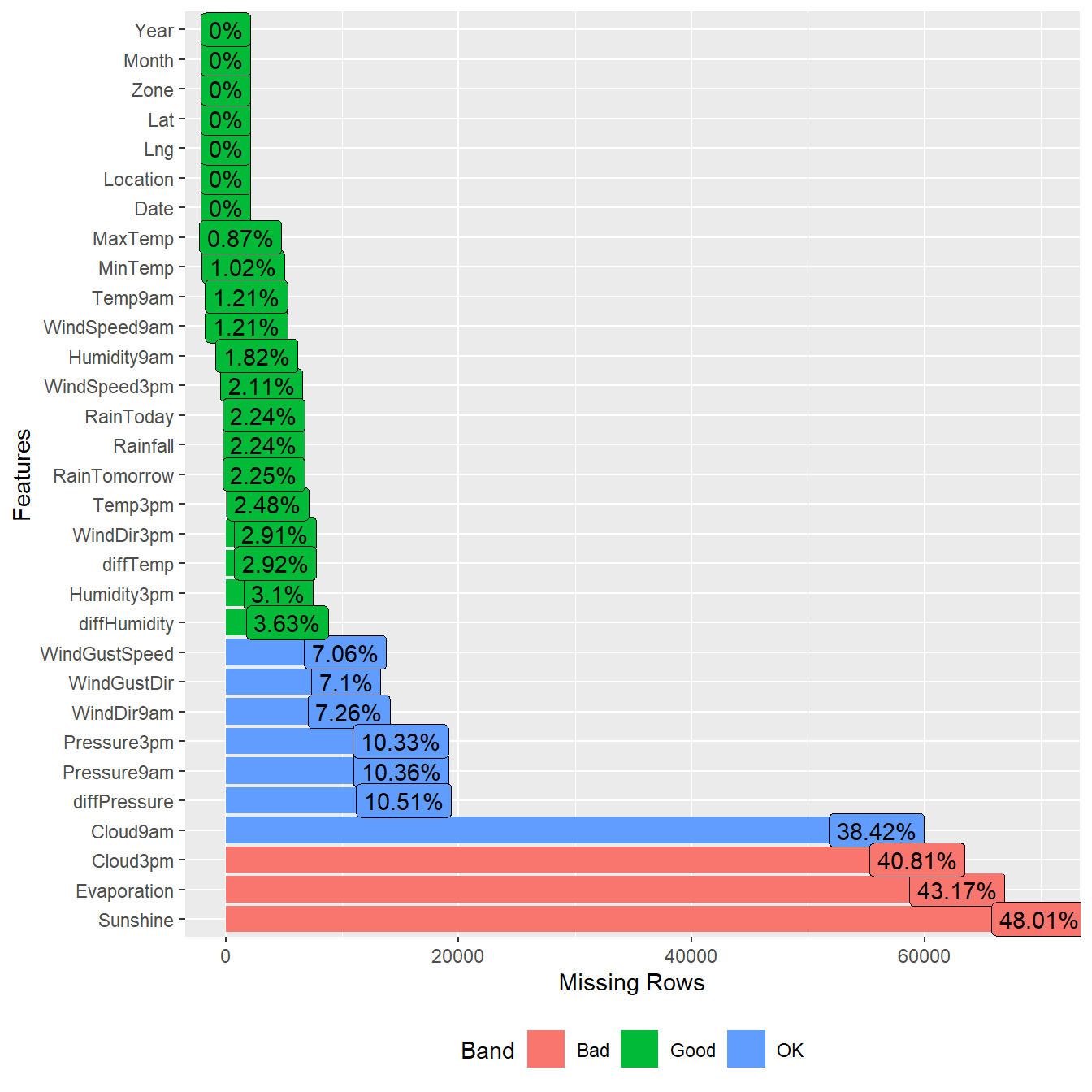
Evaluate Missing Values
Let’s examine the data for missing values.
# Inspect missing data using DataExplorer
DataExplorer::plot_missing(dat)
Inspect Numerical Variables
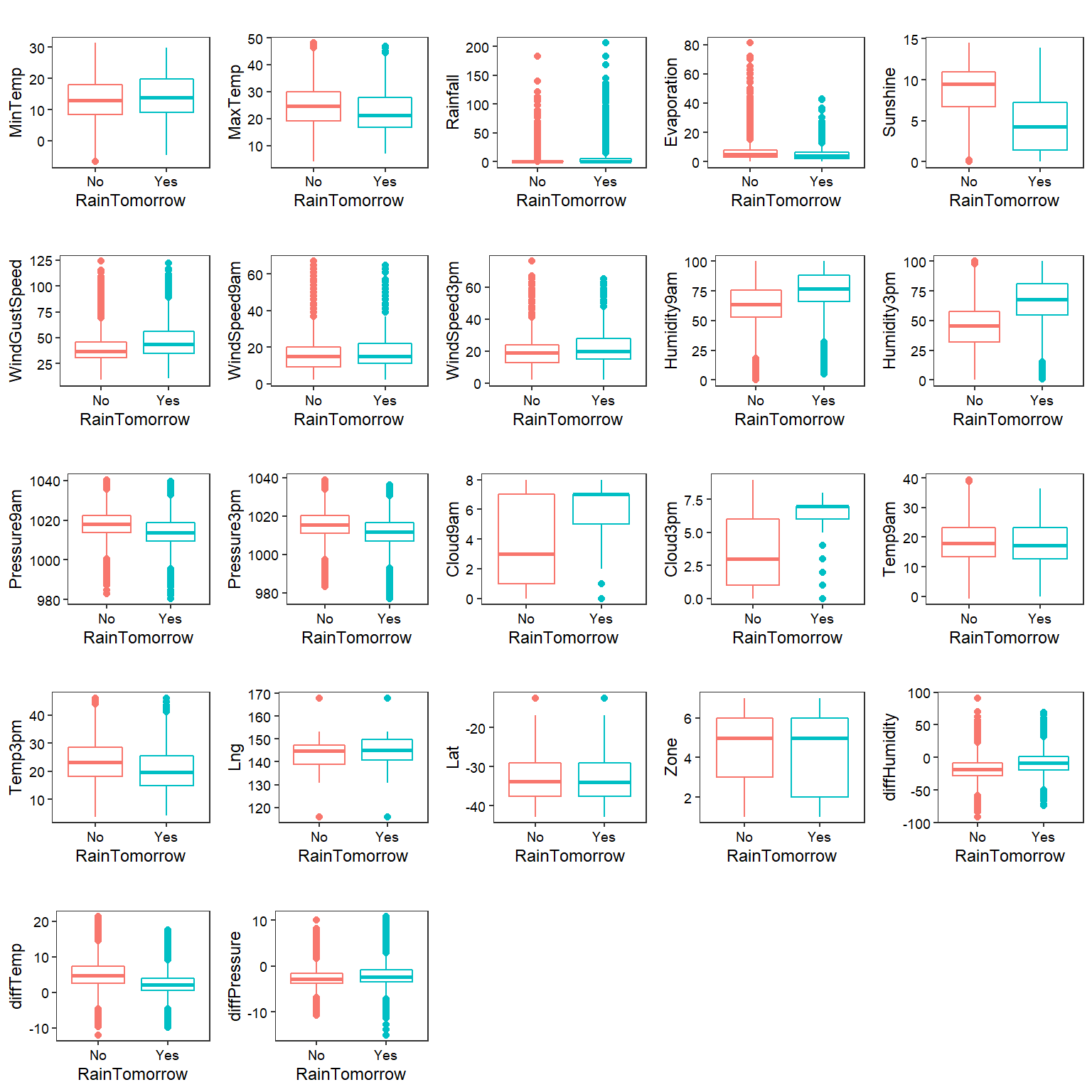
Let’s conduct some exploratory data analysis visualize, investigate, and summarize the data. First, we will inspect the numerical variables and then the categorical variables.
Box Plots
# Create box plots
plotboxes <- function(yvar) {
ggplot(data = na.omit(dat),
aes_(x = ~RainTomorrow, y = as.name(yvar), color = ~RainTomorrow)) +
geom_boxplot() +
labs(title = '',
x = 'RainTomorrow',
y = yvar) +
theme_bw(base_size = 9) +
theme(panel.grid = element_blank(),
axis.text = element_text(color = 'black'),
legend.position = 'none')
}
numeric_vars <- dat %>%
select_if(is.numeric) %>%
names()
plots <- lapply(numeric_vars, plotboxes)
cowplot::plot_grid(plotlist = plots)
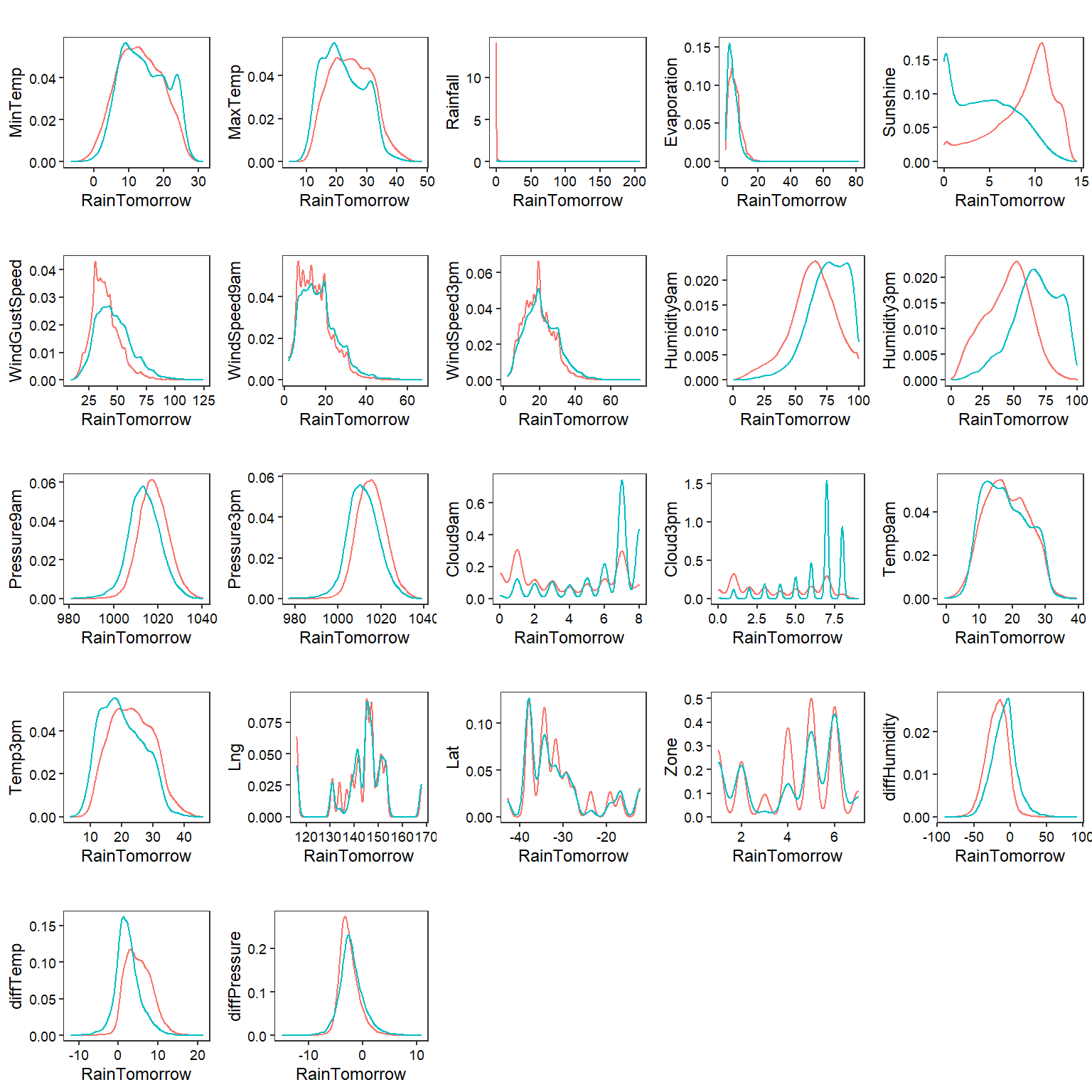
Density Plots
# Create density plots
plotdensity <- function(yvar) {
ggplot(data = na.omit(dat),
aes_(x = as.name(yvar), color = ~RainTomorrow)) +
geom_density(alpha = 0.5) +
labs(title = '',
x = 'RainTomorrow',
y = yvar) +
theme_bw(base_size = 9) +
theme(panel.grid = element_blank(),
axis.text = element_text(color = 'black'),
legend.position = 'none')
}
numeric_vars <- dat %>% select_if(is.numeric) %>% names()
plots <- lapply(numeric_vars, plotdensity)
cowplot::plot_grid(plotlist = plots)
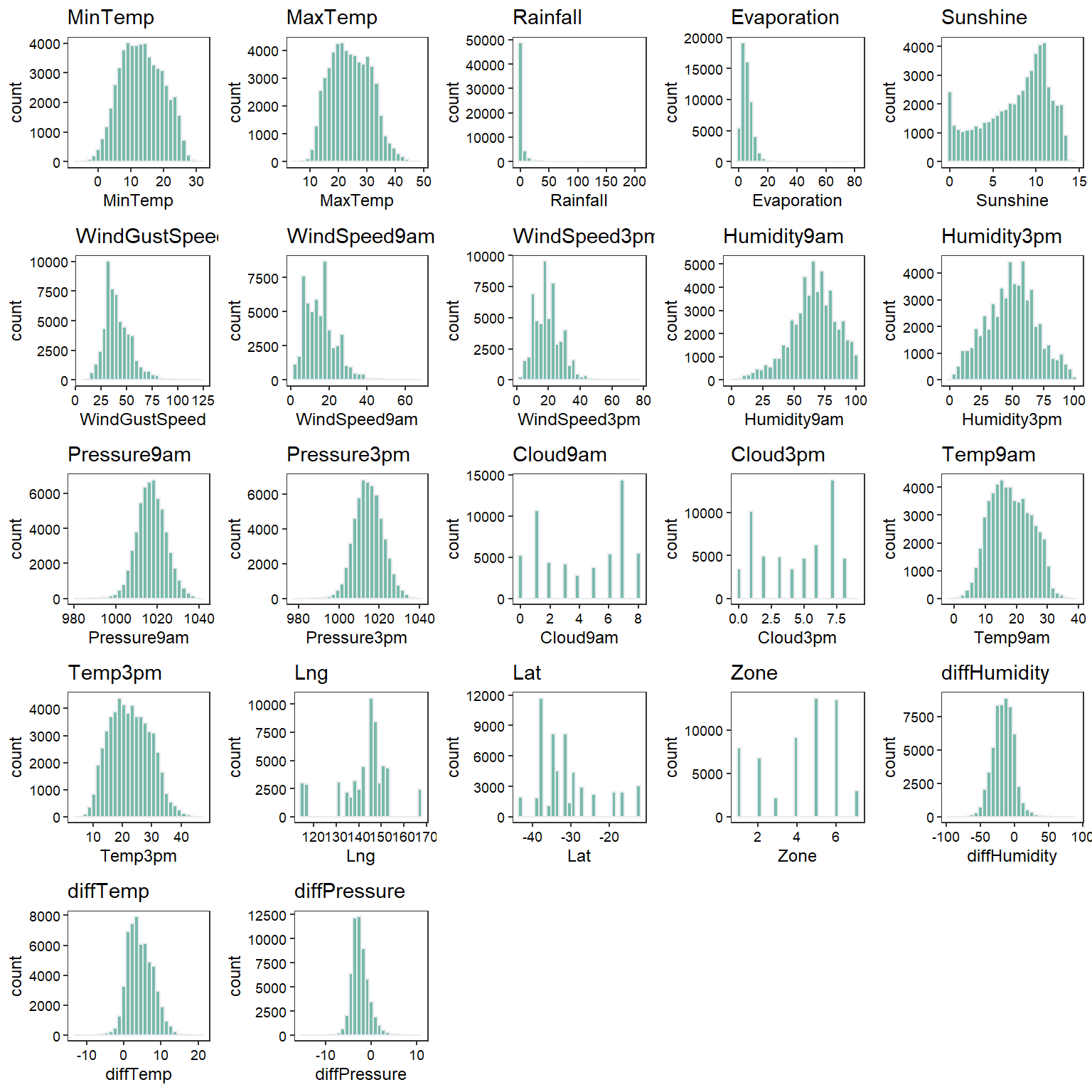
Histograms
# Create histograms
plothist <- function(yvar) {
ggplot(data = na.omit(dat), aes_(x = as.name(yvar))) +
geom_histogram(fill = '#69b3a2', color = '#e9ecef', alpha = 0.9) +
labs(title = yvar) +
theme_bw(base_size = 9) +
theme(panel.grid = element_blank(),
axis.text = element_text(color = 'black'),
legend.position = 'none')
}
plots <- lapply(numeric_vars, plothist)
cowplot::plot_grid(plotlist = plots)
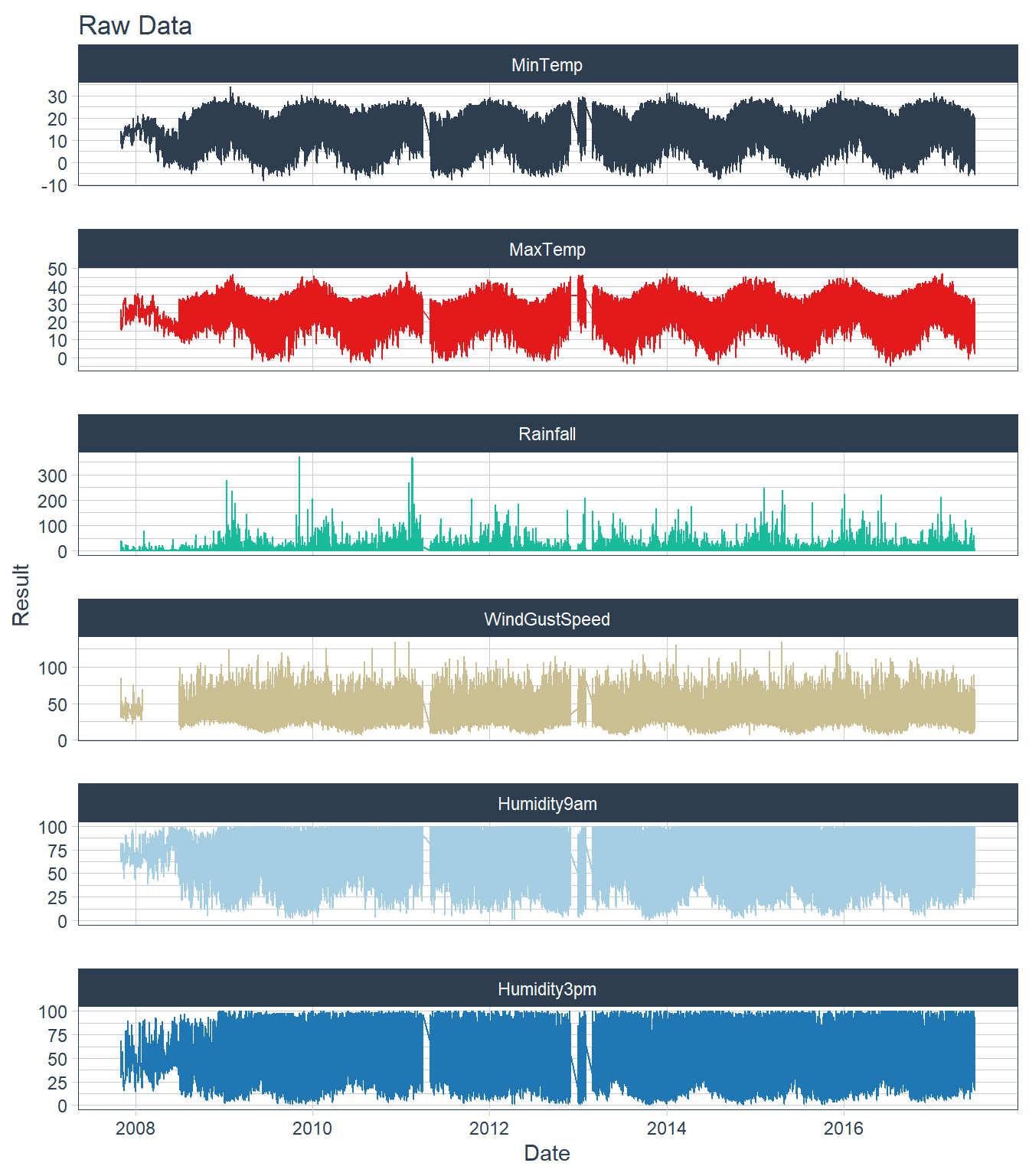
Time-Series Plots
# Convert data from wide to long
dat_long <- dat %>%
# select_if(~ is.numeric(.) | inherits(., "Date")) %>%
select(Date, MinTemp, MaxTemp, Rainfall, WindGustSpeed, Humidity9am, Humidity3pm) %>%
melt(id.vars = 'Date') %>%
setNames(., c("Date", "Param", "Result"))
# Plot dat_long using tidyquant theme
ggplot(dat_long, aes(x = Date, y = Result, color = Param)) +
geom_line() +
facet_wrap(~ Param, ncol = 1, scale = 'free_y') +
ggtitle('Raw Data') +
scale_color_tq() +
theme_tq() +
theme(legend.position = 'none')
Annual Rainfall
# Plot rainfall data by year
dat_long %>%
dplyr::filter(Param == 'Rainfall') %>%
mutate(Year = as.factor(lubridate::year(Date))) %>%
droplevels() %>%
na.omit() %>%
ggplot(aes(x = Year, y = log10(Result))) +
geom_boxplot(fill = '#69b3a2', alpha = 0.9) +
ggtitle('Rainfall') +
theme_bw(base_size = 11) +
theme(axis.text = element_text(color = 'black'))
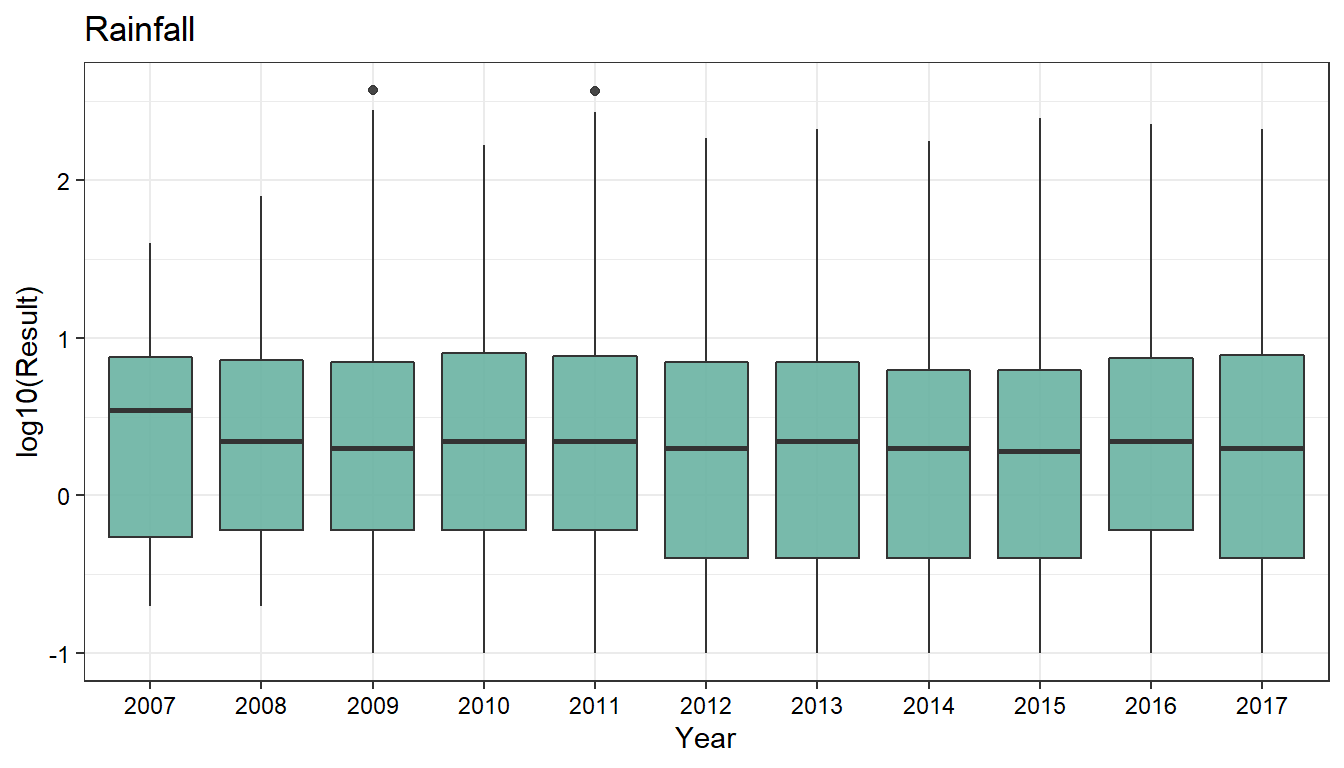
Monthly Rainfall
# Plot rainfall data by month
dat_long %>%
dplyr::filter(Param == 'Rainfall') %>%
mutate(Month = lubridate::month(Date, label = TRUE)) %>%
droplevels() %>%
na.omit() %>%
ggplot(aes(x = Month, y = log10(Result))) +
geom_boxplot(fill = '#69b3a2', alpha = 0.9) +
ggtitle('Rainfall') +
theme_bw(base_size = 11) +
theme(axis.text = element_text(color = 'black'))
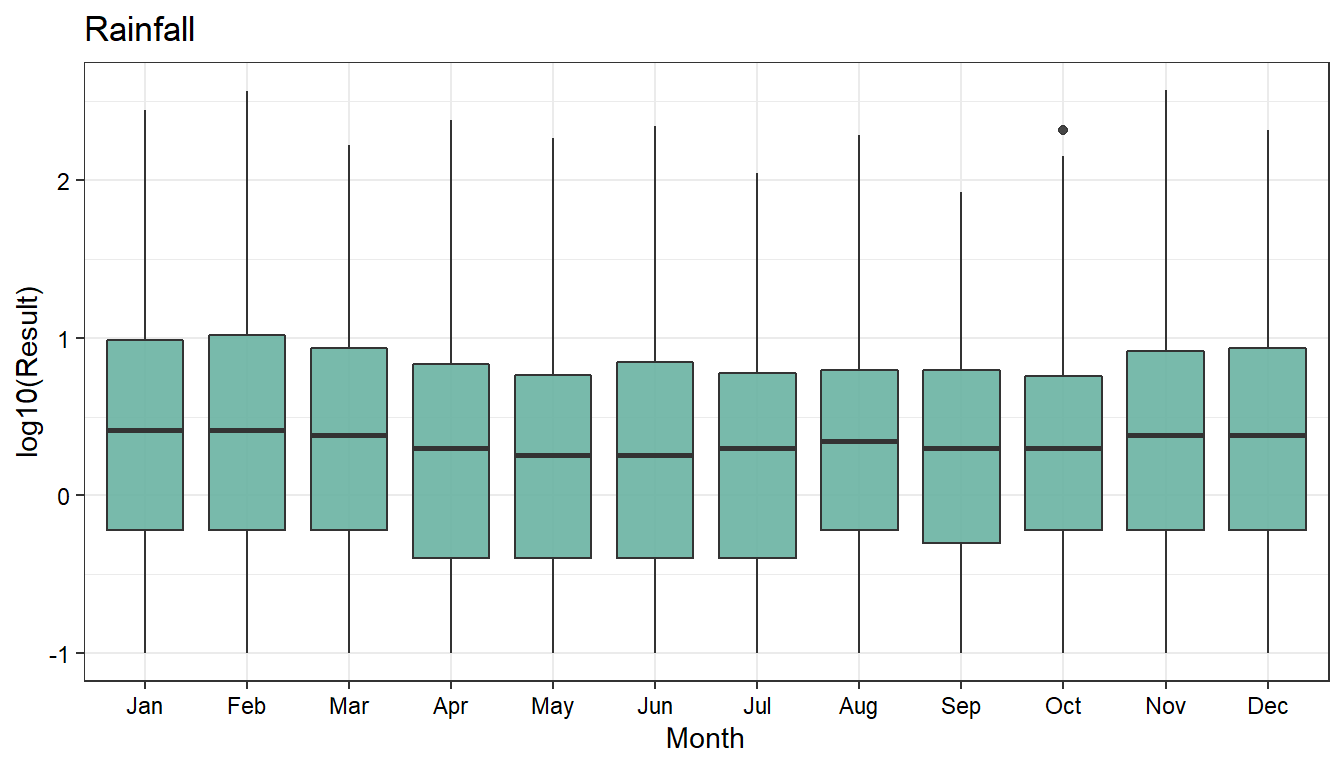
Inspect Categorical Variables
Inspect Rain by Location
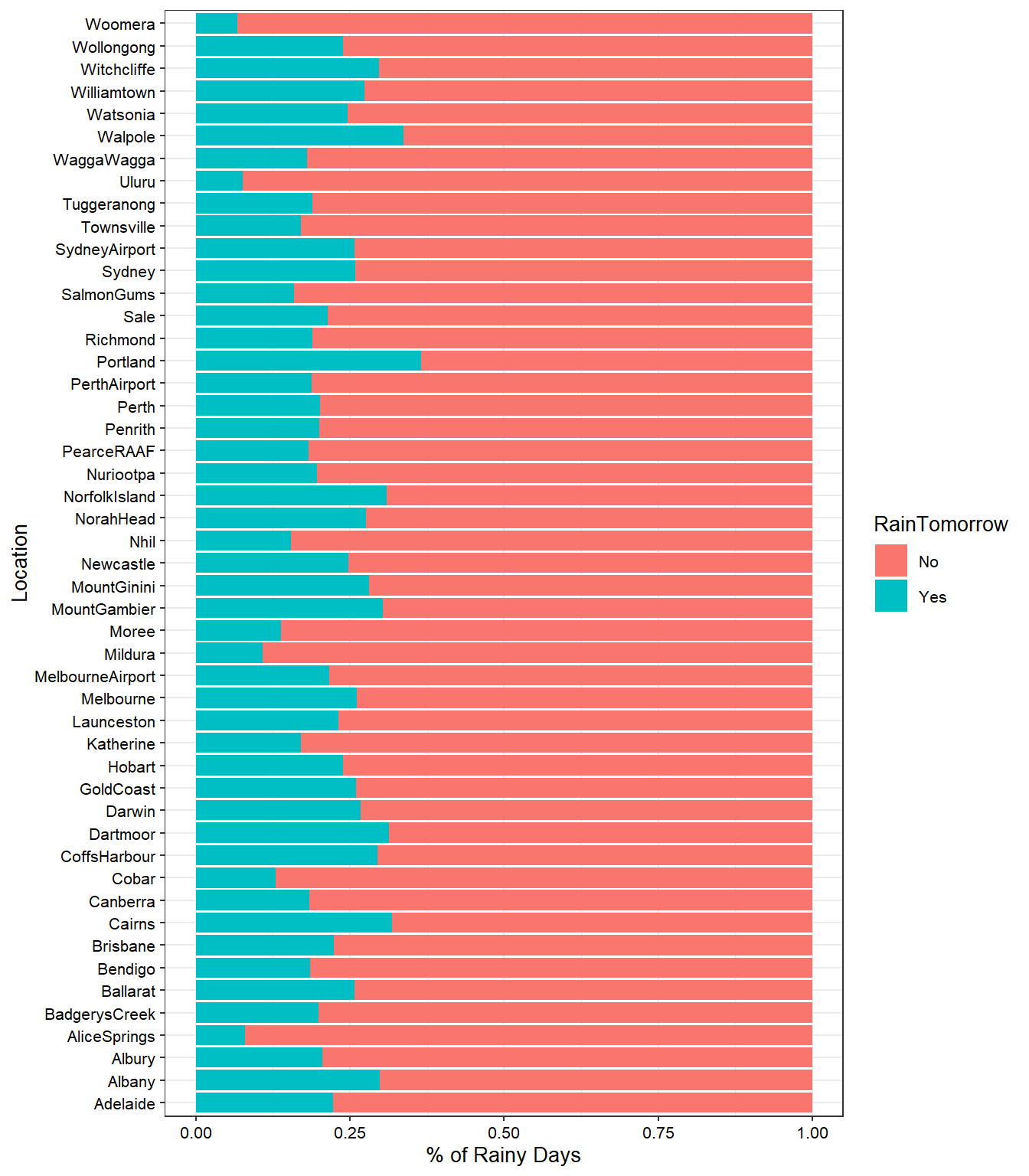
# Exploring Rain by location
dat %>%
select(Location, RainTomorrow) %>%
count(Location, RainTomorrow) %>%
drop_na() %>%
group_by(Location) %>%
mutate(percent_Rain = n / sum(n)) %>%
ggplot(aes(percent_Rain, Location, fill = RainTomorrow)) +
geom_col() +
ylab('Location') +
xlab('% of Rainy Days') +
theme_bw(base_size = 10) +
theme(axis.text = element_text(color = 'black'))
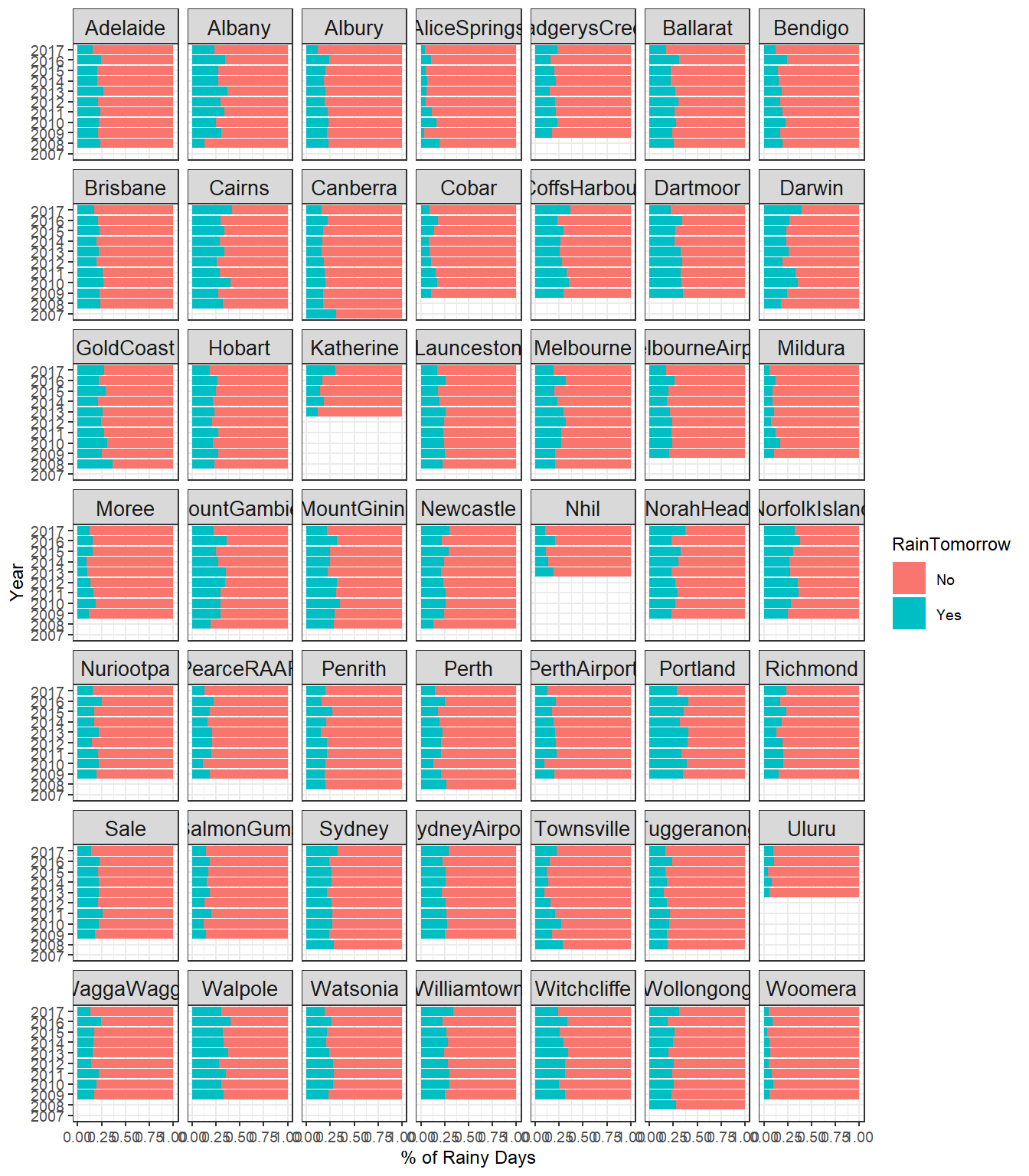
Annual Rainfall by Location
# Plot annual rainfall by location
dat %>%
count(Location, Year, RainTomorrow) %>%
drop_na() %>%
group_by(Location, Year) %>%
mutate(percent_Rain = n / sum(n)) %>%
ggplot(aes(percent_Rain , Year, fill = RainTomorrow)) +
geom_col() +
xlab('% of Rainy Days') +
theme_bw(base_size = 9) +
theme(strip.text = element_text(size = 10)) +
facet_wrap(~Location)
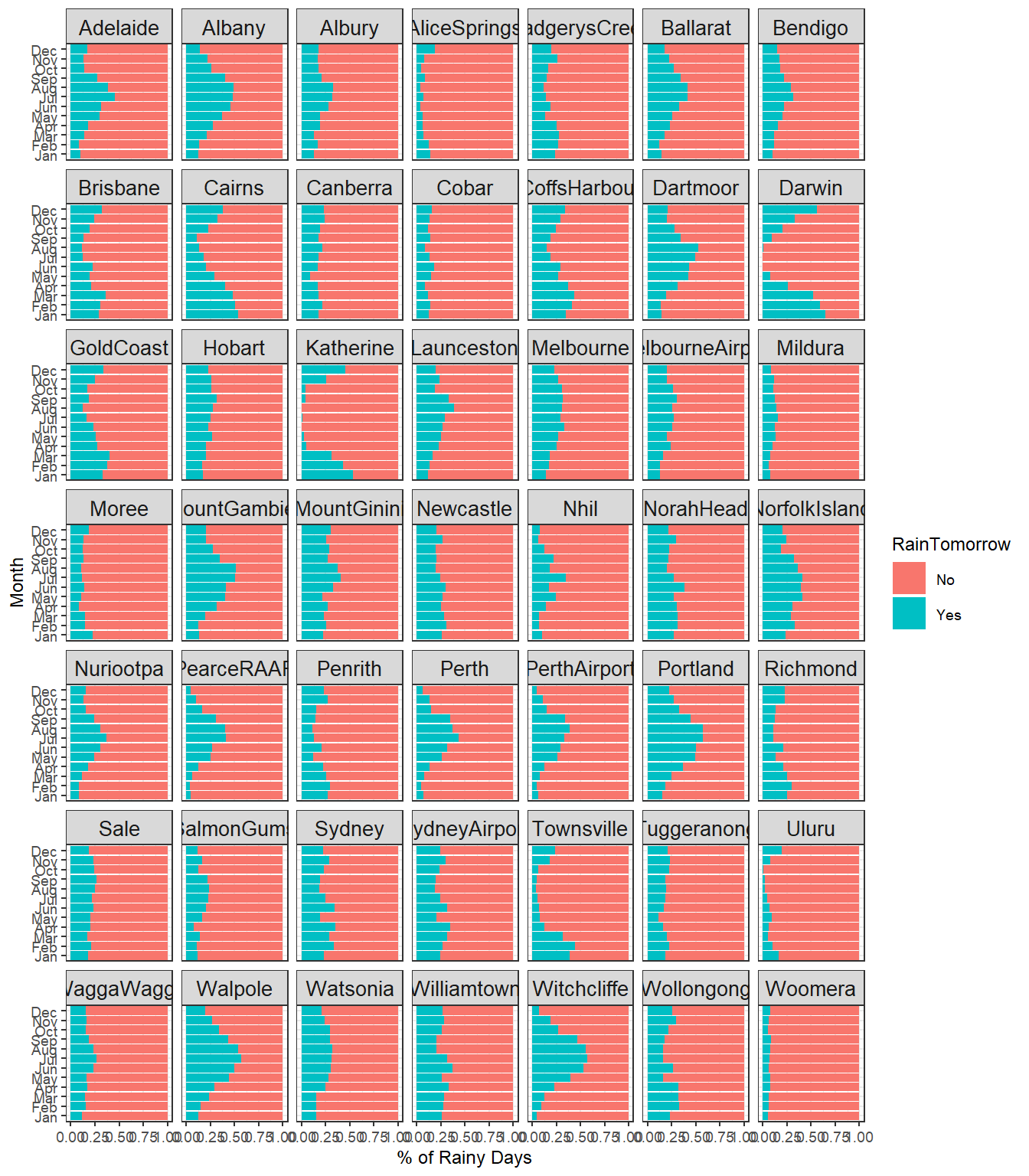
Monthly Rainfall by Location
# Plot monthly rainfall by location
dat %>%
count(Location, Month, RainTomorrow) %>%
drop_na() %>%
group_by(Location, Month) %>%
mutate(percent_Rain = n / sum(n)) %>%
ggplot(aes(percent_Rain , Month, fill = RainTomorrow)) +
geom_col() +
xlab('% of Rainy Days') +
theme_bw(base_size = 9) +
theme(strip.text = element_text(size = 10)) +
facet_wrap(~Location)
Chi-Square Test
Let’s calculate Chi-square between each feature and the target variable to determine the dependence/independence of the variables.
# Chi-Square to check whether the variables are dependent on RainTomorrow
factor_vars <- names(dat)[!is.element(names(dat), c(numeric_vars, 'RainTomorrow'))]
factor_vars
## [1] "Date" "Location" "WindGustDir" "WindDir9am" "WindDir3pm"
## [6] "RainToday" "Month" "Year"
chisq_test_res <- lapply(factor_vars, function(x) {
chisq.test(dat[,x], dat[, 'RainTomorrow'], simulate.p.value = TRUE)
})
names(chisq_test_res) <- factor_vars
chisq_test_res
## $Date
##
## Pearson's Chi-squared test with simulated p-value (based on 2000
## replicates)
##
## data: dat[, x] and dat[, "RainTomorrow"]
## X-squared = 16786, df = NA, p-value = 0.0004998
##
##
## $Location
##
## Pearson's Chi-squared test with simulated p-value (based on 2000
## replicates)
##
## data: dat[, x] and dat[, "RainTomorrow"]
## X-squared = 3544.8, df = NA, p-value = 0.0004998
##
##
## $WindGustDir
##
## Pearson's Chi-squared test with simulated p-value (based on 2000
## replicates)
##
## data: dat[, x] and dat[, "RainTomorrow"]
## X-squared = 1519.9, df = NA, p-value = 0.0004998
##
##
## $WindDir9am
##
## Pearson's Chi-squared test with simulated p-value (based on 2000
## replicates)
##
## data: dat[, x] and dat[, "RainTomorrow"]
## X-squared = 2214.8, df = NA, p-value = 0.0004998
##
##
## $WindDir3pm
##
## Pearson's Chi-squared test with simulated p-value (based on 2000
## replicates)
##
## data: dat[, x] and dat[, "RainTomorrow"]
## X-squared = 1281.3, df = NA, p-value = 0.0004998
##
##
## $RainToday
##
## Pearson's Chi-squared test with simulated p-value (based on 2000
## replicates)
##
## data: dat[, x] and dat[, "RainTomorrow"]
## X-squared = 13801, df = NA, p-value = 0.0004998
##
##
## $Month
##
## Pearson's Chi-squared test with simulated p-value (based on 2000
## replicates)
##
## data: dat[, x] and dat[, "RainTomorrow"]
## X-squared = 469.53, df = NA, p-value = 0.0004998
##
##
## $Year
##
## Pearson's Chi-squared test with simulated p-value (based on 2000
## replicates)
##
## data: dat[, x] and dat[, "RainTomorrow"]
## X-squared = 184.2, df = NA, p-value = 0.0004998
Spearman Correlation
Let’s measure the strength of the relationship between variables. We will use label encoding for the categorical variables.
# Compute a correlation matrix
method <- 'spearman'
main_title <- ifelse(
method == 'spearman', 'Spearman Correlation', 'Pearson Correlation'
)
# Create heat map of correlation matrix with formatting
dat %>%
drop_na() %>%
mutate(across(where(is.character), as.factor)) %>%
mutate(across(where(is.factor), as.integer)) %>%
dplyr::select(-Date) %>%
cor(., method = method) %>%
ggcorrplot(., hc.order = TRUE, type = "lower",
outline.col = 'white',
ggtheme = ggplot2::theme_gray,
colors = c('#6D9EC1', 'white', '#E46726'),
lab = TRUE, lab_size = 2.5) +
labs(title = main_title) +
theme(plot.title = element_text(margin = margin(b = 3), size = 14,
hjust = 0.0, color = 'black', face = 'bold'),
axis.text.x = element_text(size = 9, angle = 90, color = 'black'),
axis.text.y = element_text(size = 9, color = 'black'))
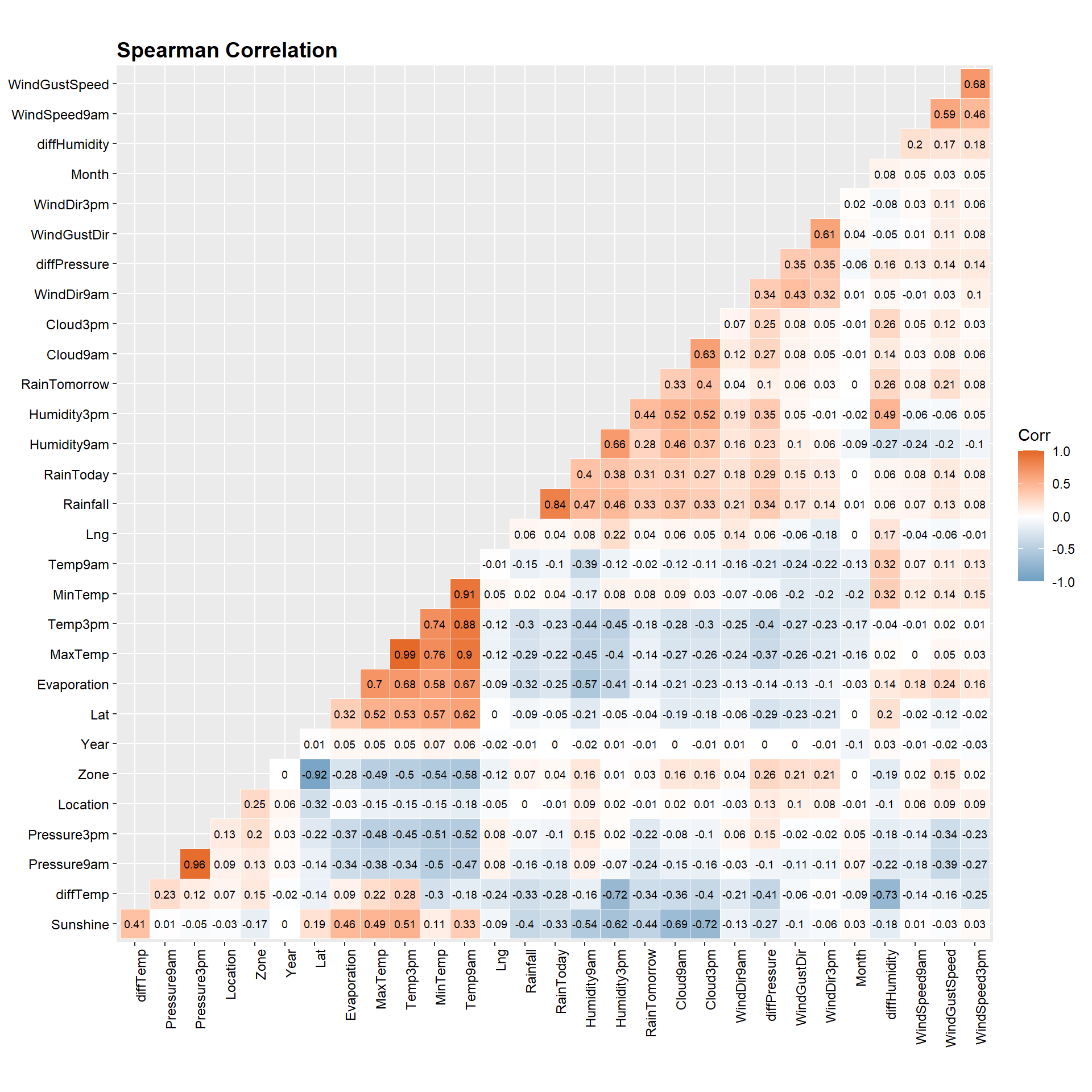
Let’s get a sorted list of the correlation between each feature and the target variable RainTomorrow.
# Get correlation with RainTomorrow
dat %>%
# dplyr::select(-c(Cloud9am, Cloud3pm, Evaporation, Sunshine)) %>%
drop_na() %>%
mutate(across(where(is.character), as.factor)) %>%
mutate(across(where(is.factor), as.integer)) %>%
dplyr::select(-Date) %>%
cor(., method = method) %>%
round(., 2) %>%
as.table() %>%
data.frame() %>%
filter(Var2 == 'RainTomorrow') %>%
dplyr::select(-Var2) %>%
arrange(desc(Freq))
## Var1 Freq
## 1 RainTomorrow 1.00
## 2 Humidity3pm 0.44
## 3 Cloud3pm 0.40
## 4 Rainfall 0.33
## 5 Cloud9am 0.33
## 6 RainToday 0.31
## 7 Humidity9am 0.28
## 8 diffHumidity 0.26
## 9 WindGustSpeed 0.21
## 10 diffPressure 0.10
## 11 MinTemp 0.08
## 12 WindSpeed9am 0.08
## 13 WindSpeed3pm 0.08
## 14 WindGustDir 0.06
## 15 WindDir9am 0.04
## 16 Lng 0.04
## 17 WindDir3pm 0.03
## 18 Zone 0.03
## 19 Month 0.00
## 20 Location -0.01
## 21 Year -0.01
## 22 Temp9am -0.02
## 23 Lat -0.04
## 24 MaxTemp -0.14
## 25 Evaporation -0.14
## 26 Temp3pm -0.18
## 27 Pressure3pm -0.22
## 28 Pressure9am -0.24
## 29 diffTemp -0.34
## 30 Sunshine -0.44
Final Data Set
Here we select the variables that have some relation with the target variable RainTomorrow. This new dataset with a reduced number of variables will be used for building our classification models. Although Sunshine contains a large fraction of missing values, it is strongly correlated with RainTomorrow.
# Prep final data
dat2 <- dat %>%
dplyr::select(
RainTomorrow,
RainToday,
Zone,
Month,
Sunshine,
diffTemp,
diffPressure,
diffHumidity,
WindGustSpeed,
MaxTemp,
Temp3pm ,
Rainfall,
Humidity9am,
Humidity3pm,
Pressure9am,
Pressure3pm
) %>%
tidyr::drop_na() %>%
droplevels() %>%
mutate_if(is.character, as.factor) %>%
mutate(
Month = factor(Month, ordered = FALSE),
RainTomorrow = relevel(RainTomorrow, ref = 'Yes')
)
# Check structure of final data
str(dat2)
## 'data.frame': 69124 obs. of 16 variables:
## $ RainTomorrow : Factor w/ 2 levels "Yes","No": 2 2 2 2 2 2 2 2 2 2 ...
## $ RainToday : Factor w/ 2 levels "No","Yes": 1 1 1 1 1 1 1 1 1 1 ...
## $ Zone : int 4 4 4 4 4 4 4 4 4 4 ...
## $ Month : Factor w/ 12 levels "Jan","Feb","Mar",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ Sunshine : num 12.3 13 10.6 12.2 8.4 0 12.6 13.2 12.3 12.7 ...
## $ diffTemp : num 6.8 6.7 6.2 6.5 4 3.6 6.5 12.1 9.9 9.6 ...
## $ diffPressure : num -1.9 -0.8 -3.1 -3.6 -3.3 ...
## $ diffHumidity : int -7 -22 -20 -15 -4 -7 -18 -16 -18 -47 ...
## $ WindGustSpeed: int 48 37 46 31 35 43 41 37 48 41 ...
## $ MaxTemp : num 35.2 28.9 37.6 38.4 41 36.1 34 34.2 35.5 35.5 ...
## $ Temp3pm : num 33.4 27 34.9 35.6 37.6 34.3 31.5 32.8 33.3 33.6 ...
## $ Rainfall : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Humidity9am : int 20 30 42 37 19 26 33 25 46 61 ...
## $ Humidity3pm : int 13 8 22 22 15 19 15 9 28 14 ...
## $ Pressure9am : num 1006 1013 1012 1013 1011 ...
## $ Pressure3pm : num 1004 1012 1009 1009 1007 ...
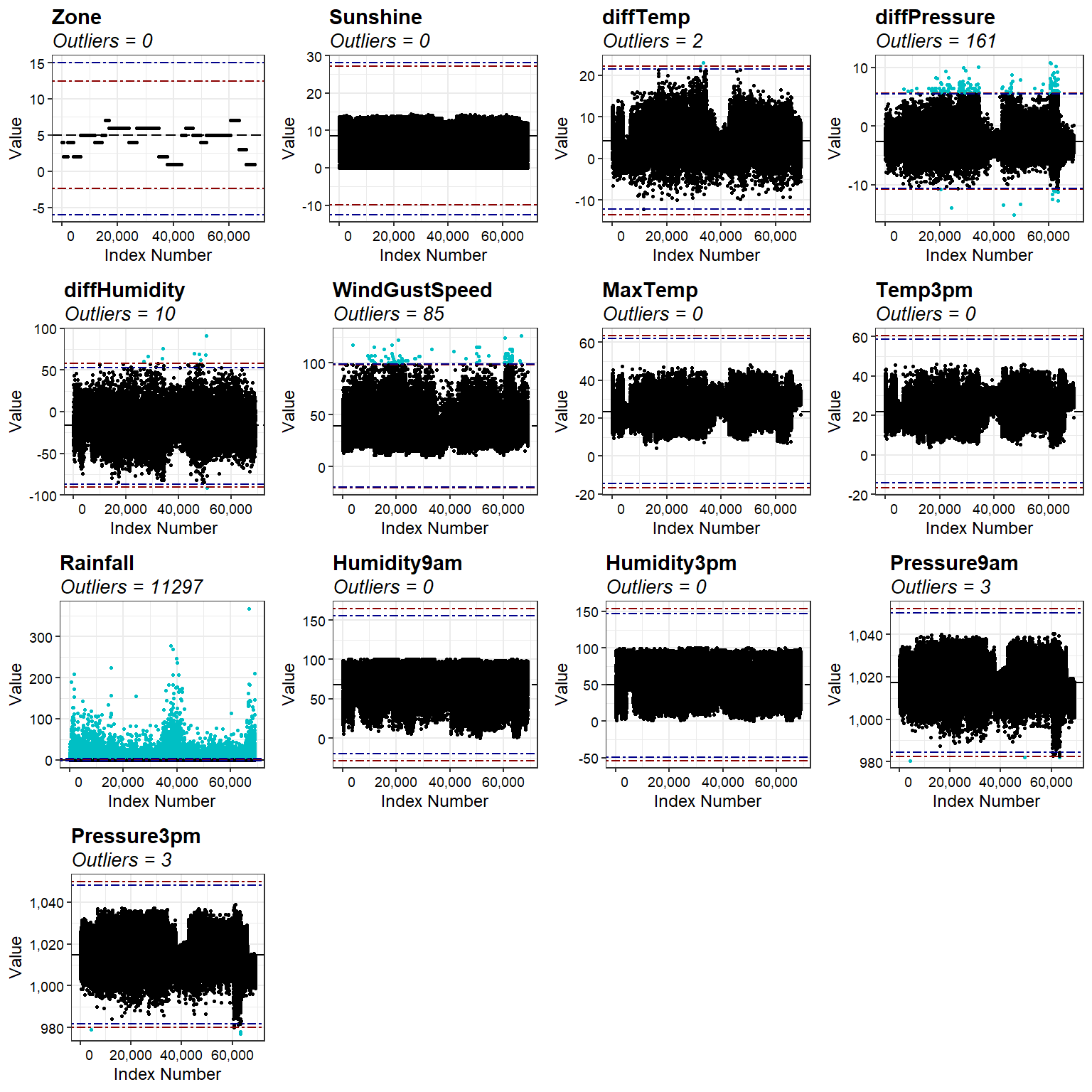
Outlier Evaluation
Let’s create outlier plots using the interquartile range (IQR) score and the median of the absolute deviations from the median (MAD) score. For this analysis, observations are identified as outliers if their value exceeds three times the IQR score and five times the MAD score. Because statistical outliers may actually be part of the normal population of the data, we will not remove or modify them.
# Create outlier plots
numeric_vars <- dat2 %>% select_if(is.numeric) %>% names()
data_outliers <- lapply(numeric_vars, find_outliers, d = dat2)
plots <- lapply(data_outliers, outlier_plots)
cowplot::plot_grid(plotlist = plots)
Scale Variables
Let’s scale the numeric variables. Tree-based ensemble methods and logistic regression for classification are invariant to feature scaling, but scaling is helpful for gradient descent-based algorithms, such as neutral networks.
# Rescale numeric variables
dat2 <- dat2 %>%
mutate(across(Sunshine:Pressure3pm, scales::rescale))
Model Setup
Data Splitting
Let’s split the data into training and testing datasets within the stratification variable RainTomorrow. This helps ensure that the splits have equivalent proportions as the original data. We will tune each model’s hyperparameters using a grid search with 5-fold cross-validation.
# Load libraries
library(tidymodels)
# Split data in training and testing sets
set.seed(123)
dat2_split <- initial_split(dat2, prop = 0.75, strata = RainTomorrow)
dat2_train <- training(dat2_split)
dat2_test <- testing(dat2_split)
# Create cross-validation data sets
dat2_cv <- vfold_cv(dat2_train, v = 5, strata = RainTomorrow)
Model Specification
We will build the following classification models using the tidymodels framework:
- Logistic Regression
- Random Forest
- XGBoost (extreme gradient boosted trees)
- Neural network
First, let’s build specifications for each model. In this step we specify the models, the engine to fit the model, and other specifications.
# Logistic Regression
glm_spec <- logistic_reg() %>%
set_engine('glm')
glm_spec
## Logistic Regression Model Specification (classification)
##
## Computational engine: glm
# XGBoost
xgb_spec <- boost_tree(
trees = 500,
tree_depth = tune(), min_n = tune(),
loss_reduction = tune(), # first three: model complexity
sample_size = tune(), mtry = tune(), # randomness
learn_rate = tune(), # step size
) %>%
set_engine('xgboost') %>%
set_mode('classification')
xgb_spec
## Boosted Tree Model Specification (classification)
##
## Main Arguments:
## mtry = tune()
## trees = 500
## min_n = tune()
## tree_depth = tune()
## learn_rate = tune()
## loss_reduction = tune()
## sample_size = tune()
##
## Computational engine: xgboost
# Random Forest
rf_spec <- rand_forest(
mtry = tune(), # tune via cross-validation
trees = 500, # grow 500 random trees
min_n = tune() # tune via cross-validation
) %>%
set_mode('classification') %>%
set_engine('ranger')
rf_spec
## Random Forest Model Specification (classification)
##
## Main Arguments:
## mtry = tune()
## trees = 500
## min_n = tune()
##
## Computational engine: ranger
# Neural Network
mlp_spec <-
mlp(hidden_units = tune(), penalty = tune(), epochs = tune()) %>%
set_mode('classification') %>%
set_engine('nnet', MaxNWts = 84581, trace = 0)
mlp_spec
## Single Layer Neural Network Model Specification (classification)
##
## Main Arguments:
## hidden_units = tune()
## penalty = tune()
## epochs = tune()
##
## Engine-Specific Arguments:
## MaxNWts = 84581
## trace = 0
##
## Computational engine: nnet
Model Workflow
Now, let’s create a basic model workflow. In this step we specify the formula for the model. Later, we will add the model in the workflow.
# Create workflow
dat2_wf <- workflow() %>%
add_formula(RainTomorrow ~ .)
dat2_wf
## ══ Workflow ════════════════════════════════════════════════════════════════════
## Preprocessor: Formula
## Model: None
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## RainTomorrow ~ .
# Parallel processing
doParallel::registerDoParallel()
Logistic Regression Model
Logistic regression is one of the most simple machine learning models. It is an extremely popular approach used for classification tasks.
Fit logistic regression model on cross-validation data sets
# Fit logistic on cross-validation data sets
glm_rs <- dat2_wf %>%
add_model(glm_spec) %>%
fit_resamples(
resamples = dat2_cv,
control = control_resamples(save_pred = TRUE)
)
glm_rs
## # Resampling results
## # 5-fold cross-validation using stratification
## # A tibble: 5 × 5
## splits id .metrics .notes .predictions
## <list> <chr> <list> <list> <list>
## 1 <split [41472/10370]> Fold1 <tibble [2 × 4]> <tibble [1 × 3]> <tibble>
## 2 <split [41474/10368]> Fold2 <tibble [2 × 4]> <tibble [1 × 3]> <tibble>
## 3 <split [41474/10368]> Fold3 <tibble [2 × 4]> <tibble [1 × 3]> <tibble>
## 4 <split [41474/10368]> Fold4 <tibble [2 × 4]> <tibble [1 × 3]> <tibble>
## 5 <split [41474/10368]> Fold5 <tibble [2 × 4]> <tibble [1 × 3]> <tibble>
##
## There were issues with some computations:
##
## - Warning(s) x5: prediction from a rank-deficient fit may be misleading
##
## Run `show_notes(.Last.tune.result)` for more information.
# Assess performance of logistic model
collect_metrics(glm_rs)
## # A tibble: 2 × 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 accuracy binary 0.854 5 0.00108 Preprocessor1_Model1
## 2 roc_auc binary 0.884 5 0.00103 Preprocessor1_Model1
# Sensitivity and specificity of logistic model
glm_rs %>%
collect_predictions() %>%
sensitivity(RainTomorrow, .pred_class)
## # A tibble: 1 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 sensitivity binary 0.535
glm_rs %>%
collect_predictions() %>%
specificity(RainTomorrow, .pred_class)
## # A tibble: 1 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 specificity binary 0.944
# Confusion matrix of logistic model
glm_rs %>%
conf_mat_resampled()
## # A tibble: 4 × 3
## Prediction Truth Freq
## <fct> <fct> <dbl>
## 1 Yes Yes 1221.
## 2 Yes No 452.
## 3 No Yes 1063.
## 4 No No 7634.
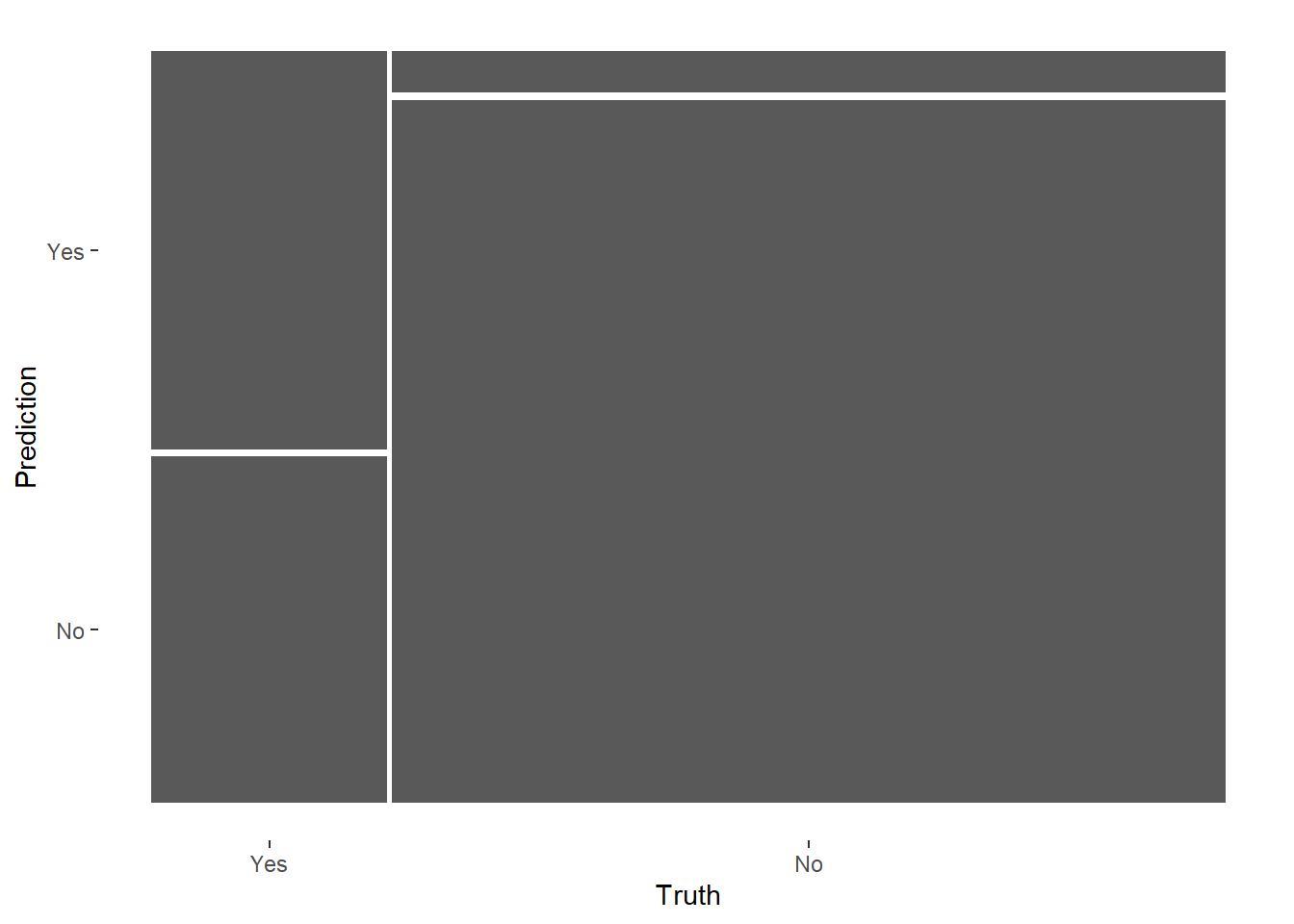
# Visualize model predictions as mosaic
glm_rs %>%
collect_predictions() %>%
conf_mat(RainTomorrow, .pred_class) %>%
autoplot(type = 'mosaic')
Final logistic regression model
# Fit logistic model on entire training data
glm_final <- dat2_wf %>%
add_model(glm_spec) %>%
last_fit(dat2_split)
glm_final
## # Resampling results
## # Manual resampling
## # A tibble: 1 × 6
## splits id .metrics .notes .predict…¹ .workflow
## <list> <chr> <list> <list> <list> <list>
## 1 <split [51842/17282]> train/test split <tibble> <tibble> <tibble> <workflow>
## # … with abbreviated variable name ¹.predictions
##
## There were issues with some computations:
##
## - Warning(s) x1: prediction from a rank-deficient fit may be misleading
##
## Run `show_notes(.Last.tune.result)` for more information.
# Logistic model performance on test data
glm_final %>% collect_metrics()
## # A tibble: 2 × 4
## .metric .estimator .estimate .config
## <chr> <chr> <dbl> <chr>
## 1 accuracy binary 0.854 Preprocessor1_Model1
## 2 roc_auc binary 0.885 Preprocessor1_Model1
# Confusion matrix on test data
glm_final %>%
collect_predictions() %>%
conf_mat(RainTomorrow, .pred_class)
## Truth
## Prediction Yes No
## Yes 2036 751
## No 1770 12725
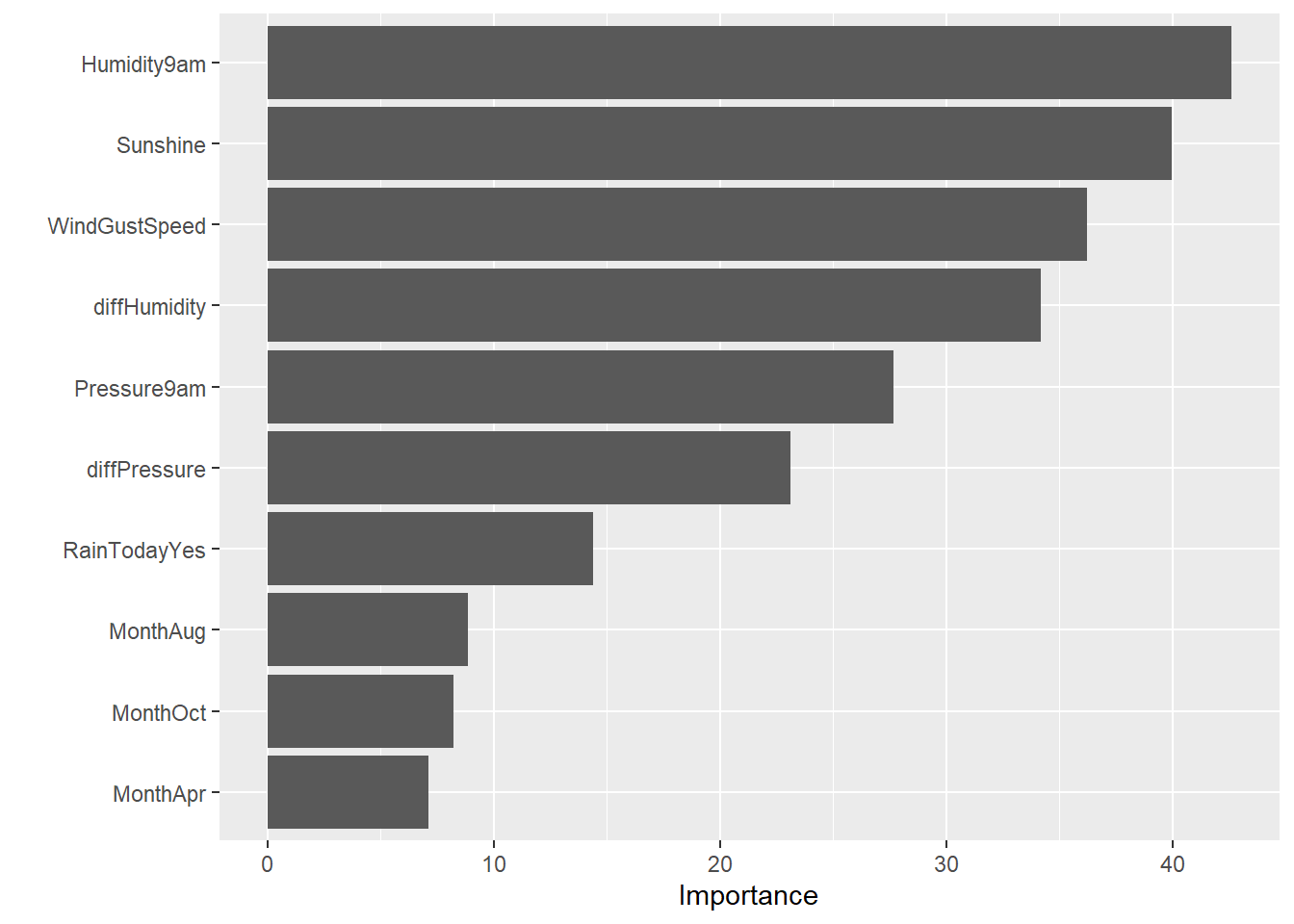
# Variable importance
glm_final %>%
extract_fit_parsnip() %>%
vip(num_features = 10)
glm_final$.workflow[[1]] %>%
tidy(exponentiate = TRUE) %>%
arrange(p.value) %>%
filter(term != "(Intercept)", p.value < 0.05)
## # A tibble: 22 × 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Sunshine 15.2 0.0681 40.0 0
## 2 Humidity9am 0.00310 0.136 -42.6 0
## 3 WindGustSpeed 0.00641 0.139 -36.2 3.49e-287
## 4 diffHumidity 0.0000238 0.312 -34.2 7.71e-256
## 5 Pressure9am 61.5 0.149 27.7 9.92e-169
## 6 diffPressure 168. 0.222 23.1 3.53e-118
## 7 RainTodayYes 0.591 0.0365 -14.4 5.84e- 47
## 8 MonthAug 0.491 0.0802 -8.87 7.52e- 19
## 9 MonthOct 0.545 0.0737 -8.23 1.83e- 16
## 10 MonthApr 0.595 0.0729 -7.12 1.12e- 12
## # … with 12 more rows
XGBoost Model
XGBoost is a popular gradient-boosted decision tree machine learning library. It is a decision tree ensemble learning algorithm similar to random forest. Let’s tune model hyperparameters using a space-filling design so we can cover the hyperparameter space as well as possible.
# Setup possible values for hyperparameters using a space-filling design
xgb_grid <- grid_latin_hypercube(
tree_depth(),
min_n(),
loss_reduction(),
sample_size = sample_prop(),
finalize(mtry(), dat2_train),
learn_rate(),
size = 30
)
xgb_grid
## # A tibble: 30 × 6
## tree_depth min_n loss_reduction sample_size mtry learn_rate
## <int> <int> <dbl> <dbl> <int> <dbl>
## 1 6 7 1.75 0.465 9 1.89e- 9
## 2 1 4 0.0000000100 0.327 4 1.20e-10
## 3 9 38 0.000000217 0.993 13 2.28e- 6
## 4 8 9 3.74 0.155 7 1.66e- 7
## 5 10 19 0.00485 0.833 2 2.05e- 7
## 6 13 31 0.000000768 0.508 3 9.64e- 6
## 7 11 35 27.2 0.383 12 1.90e- 4
## 8 9 25 0.000000618 0.248 7 4.14e- 3
## 9 7 32 0.000119 0.627 15 3.33e- 2
## 10 9 17 0.184 0.580 11 5.00e- 7
## # … with 20 more rows
# Add the xgboost model to the workflow
xgb_wf <- dat2_wf %>%
add_model(xgb_spec)
xgb_wf
## ══ Workflow ════════════════════════════════════════════════════════════════════
## Preprocessor: Formula
## Model: boost_tree()
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## RainTomorrow ~ .
##
## ── Model ───────────────────────────────────────────────────────────────────────
## Boosted Tree Model Specification (classification)
##
## Main Arguments:
## mtry = tune()
## trees = 500
## min_n = tune()
## tree_depth = tune()
## learn_rate = tune()
## loss_reduction = tune()
## sample_size = tune()
##
## Computational engine: xgboost
Tune XGBoost Model
# Tune xgboost model
set.seed(123)
xgb_res <- tune_grid(
xgb_wf,
resamples = dat2_cv,
grid = xgb_grid,
control = control_grid(save_pred = TRUE)
)
xgb_res
## # Tuning results
## # 5-fold cross-validation using stratification
## # A tibble: 5 × 5
## splits id .metrics .notes .predictions
## <list> <chr> <list> <list> <list>
## 1 <split [41472/10370]> Fold1 <tibble [60 × 10]> <tibble [0 × 3]> <tibble>
## 2 <split [41474/10368]> Fold2 <tibble [60 × 10]> <tibble [0 × 3]> <tibble>
## 3 <split [41474/10368]> Fold3 <tibble [60 × 10]> <tibble [0 × 3]> <tibble>
## 4 <split [41474/10368]> Fold4 <tibble [60 × 10]> <tibble [0 × 3]> <tibble>
## 5 <split [41474/10368]> Fold5 <tibble [60 × 10]> <tibble [0 × 3]> <tibble>
# Assess performance of xgboost model
collect_metrics(xgb_res)
## # A tibble: 60 × 12
## mtry min_n tree_depth learn_rate loss_…¹ sampl…² .metric .esti…³ mean n
## <int> <int> <int> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <int>
## 1 9 7 6 1.89e- 9 1.75e+0 0.465 accura… binary 0.854 5
## 2 9 7 6 1.89e- 9 1.75e+0 0.465 roc_auc binary 0.879 5
## 3 4 4 1 1.20e-10 1.00e-8 0.327 accura… binary 0.220 5
## 4 4 4 1 1.20e-10 1.00e-8 0.327 roc_auc binary 0.5 5
## 5 13 38 9 2.28e- 6 2.17e-7 0.993 accura… binary 0.855 5
## 6 13 38 9 2.28e- 6 2.17e-7 0.993 roc_auc binary 0.888 5
## 7 7 9 8 1.66e- 7 3.74e+0 0.155 accura… binary 0.853 5
## 8 7 9 8 1.66e- 7 3.74e+0 0.155 roc_auc binary 0.882 5
## 9 2 19 10 2.05e- 7 4.85e-3 0.833 accura… binary 0.842 5
## 10 2 19 10 2.05e- 7 4.85e-3 0.833 roc_auc binary 0.875 5
## # … with 50 more rows, 2 more variables: std_err <dbl>, .config <chr>, and
## # abbreviated variable names ¹loss_reduction, ²sample_size, ³.estimator
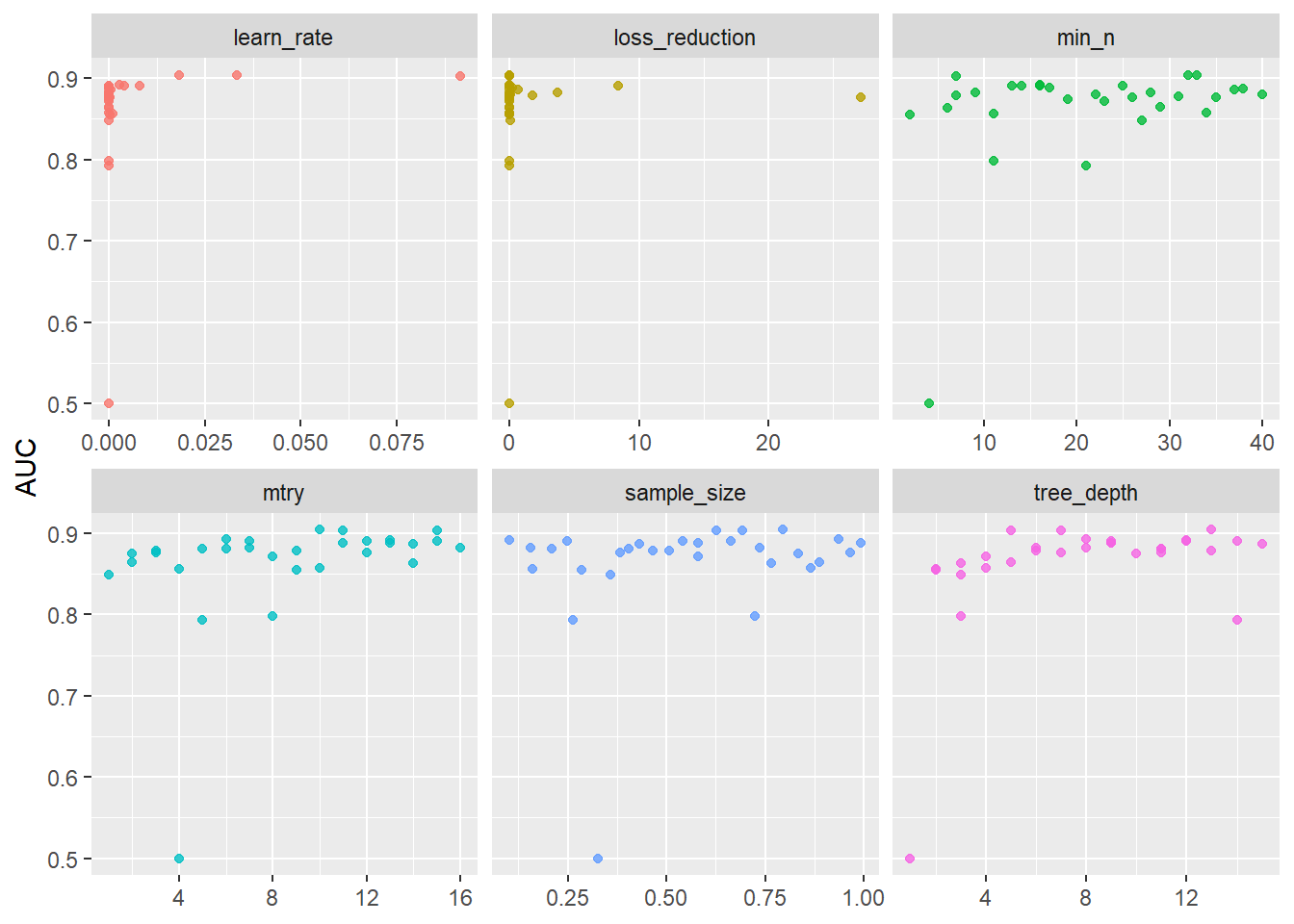
# Visualize results
xgb_res %>%
collect_metrics() %>%
filter(.metric == 'roc_auc') %>%
select(mean, mtry:sample_size) %>%
pivot_longer(mtry:sample_size,
values_to = 'value',
names_to = 'parameter'
) %>%
ggplot(aes(value, mean, color = parameter)) +
geom_point(alpha = 0.8, show.legend = FALSE) +
facet_wrap(~parameter, scales = 'free_x') +
labs(x = NULL, y = 'AUC')
Final XGBoost Model
# Show best performing sets of parameters
show_best(xgb_res, 'roc_auc')
## # A tibble: 5 × 12
## mtry min_n tree_depth learn_rate loss_r…¹ sampl…² .metric .esti…³ mean n
## <int> <int> <int> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <int>
## 1 10 33 13 0.0184 1.74e-2 0.794 roc_auc binary 0.904 5
## 2 15 32 7 0.0333 1.19e-4 0.627 roc_auc binary 0.904 5
## 3 11 7 5 0.0915 9.29e-8 0.693 roc_auc binary 0.903 5
## 4 6 16 8 0.00281 4.91e-5 0.935 roc_auc binary 0.892 5
## 5 13 16 12 0.00800 8.38e+0 0.103 roc_auc binary 0.891 5
## # … with 2 more variables: std_err <dbl>, .config <chr>, and abbreviated
## # variable names ¹loss_reduction, ²sample_size, ³.estimator
# Select best fit
best_auc <- select_best(xgb_res, 'roc_auc')
best_auc
## # A tibble: 1 × 7
## mtry min_n tree_depth learn_rate loss_reduction sample_size .config
## <int> <int> <int> <dbl> <dbl> <dbl> <chr>
## 1 10 33 13 0.0184 0.0174 0.794 Preprocessor1_Mo…
# Finalize workflow with these parameter values
final_xgb_wf <- finalize_workflow(
xgb_wf,
best_auc
)
final_xgb_wf
## ══ Workflow ════════════════════════════════════════════════════════════════════
## Preprocessor: Formula
## Model: boost_tree()
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## RainTomorrow ~ .
##
## ── Model ───────────────────────────────────────────────────────────────────────
## Boosted Tree Model Specification (classification)
##
## Main Arguments:
## mtry = 10
## trees = 500
## min_n = 33
## tree_depth = 13
## learn_rate = 0.0183655696841365
## loss_reduction = 0.01741508074499
## sample_size = 0.794431108820718
##
## Computational engine: xgboost
# Fit model on training data
xgb_final <- last_fit(final_xgb_wf, dat2_split)
# Evaluate model performance on test data
collect_metrics(xgb_final)
## # A tibble: 2 × 4
## .metric .estimator .estimate .config
## <chr> <chr> <dbl> <chr>
## 1 accuracy binary 0.867 Preprocessor1_Model1
## 2 roc_auc binary 0.904 Preprocessor1_Model1
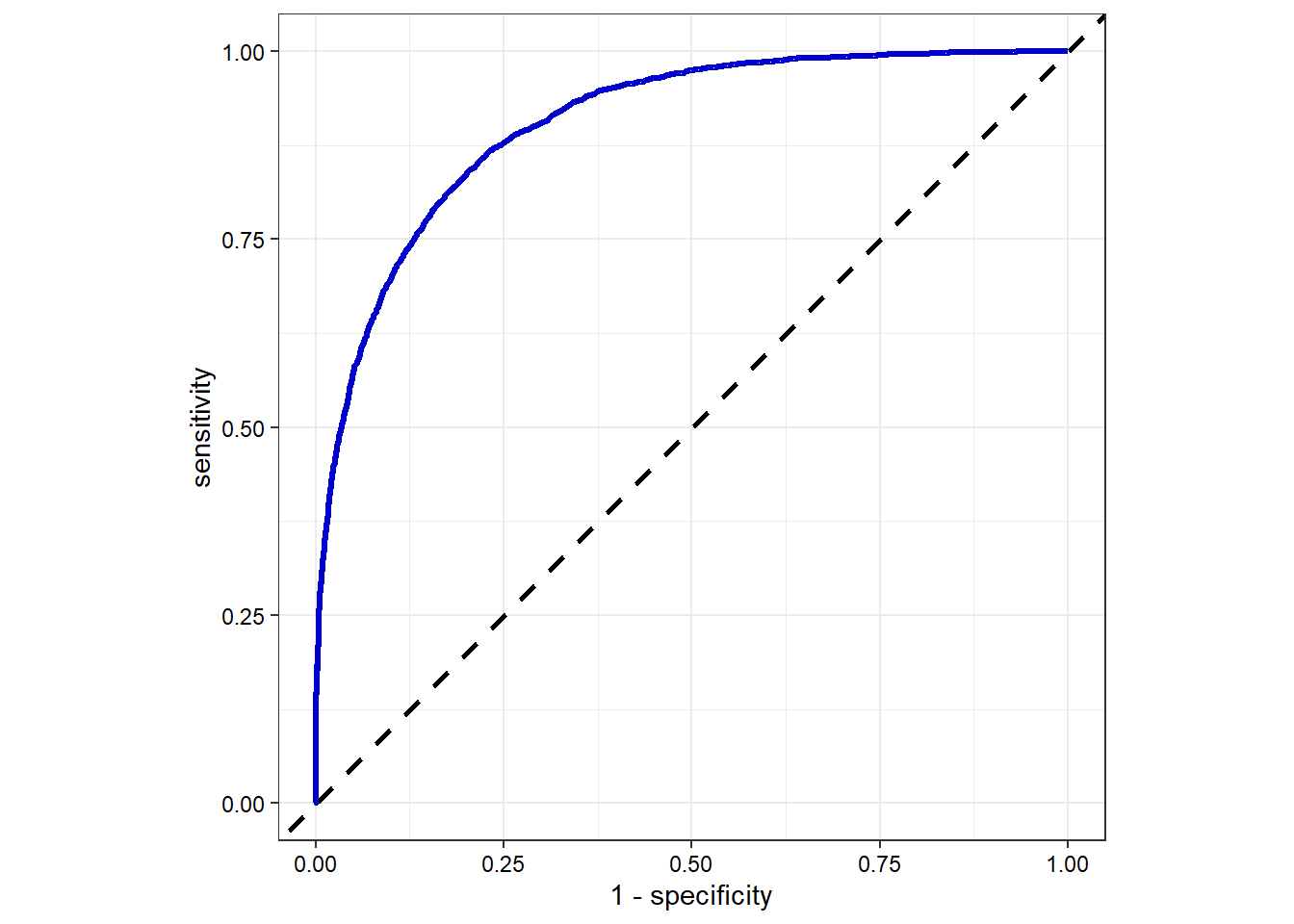
# Create ROC curve for the test data
xgb_final %>%
collect_predictions() %>%
roc_curve(RainTomorrow, .pred_Yes) %>%
ggplot(aes(x = 1 - specificity, y = sensitivity)) +
geom_line(size = 1.2, color = 'blue3') +
geom_abline(lty = 2, size = 1, color = 'black') +
coord_equal() +
theme_bw() +
theme(axis.text = element_text(color = 'black'))
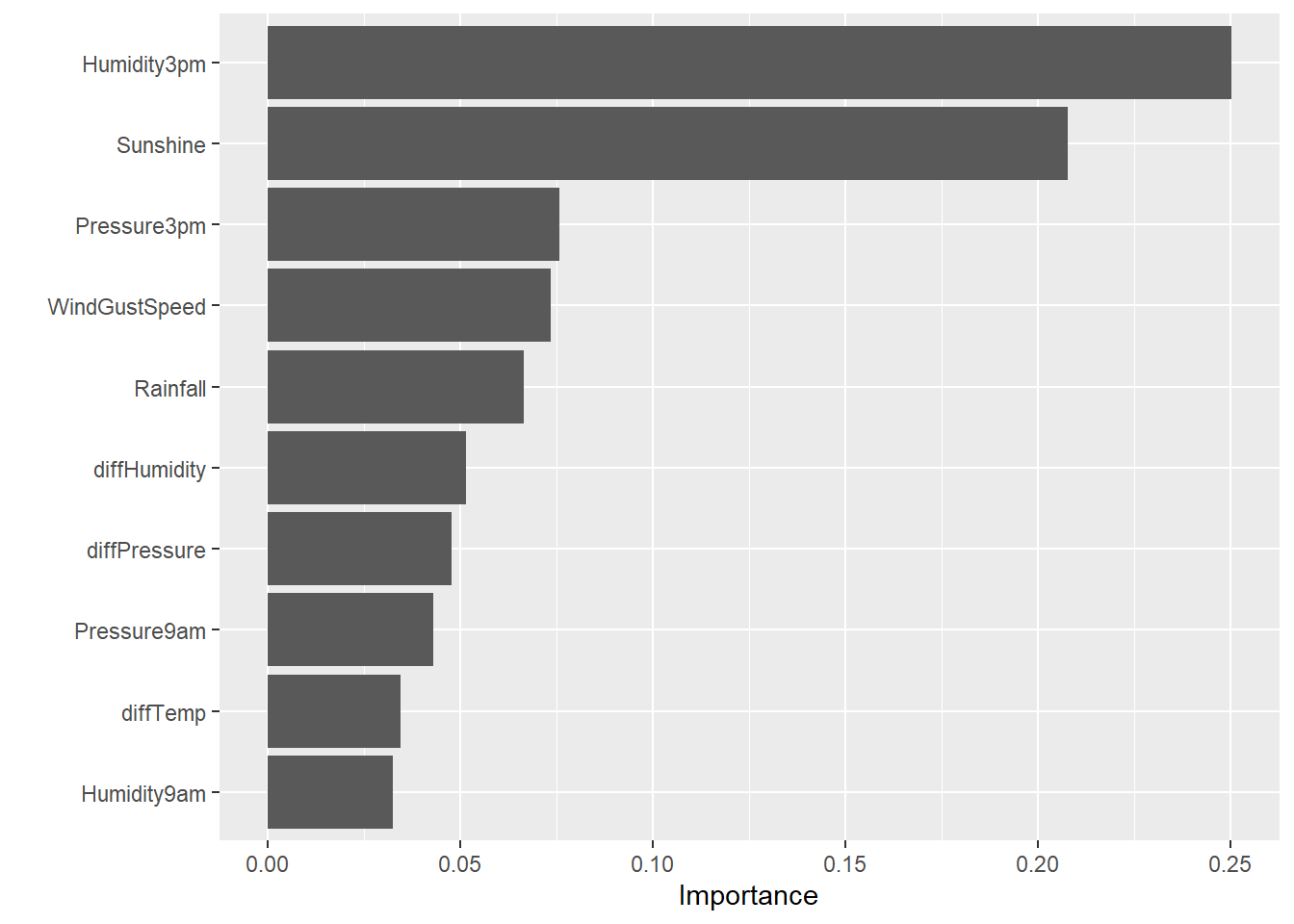
# Variable importance
final_xgb_wf %>%
fit(data = dat2_train) %>%
extract_fit_parsnip() %>%
vip(num_features = 10)
Random Forest Model
Random forest is an ensemble tree-based learning algorithm. It is commonly used due to its simplicity and diversity. Let’s use 25 combinations of mtry and min_n for tuning.
# Add the random forest model to the workflow
rf_wf <- dat2_wf %>%
add_model(rf_spec)
rf_wf
## ══ Workflow ════════════════════════════════════════════════════════════════════
## Preprocessor: Formula
## Model: rand_forest()
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## RainTomorrow ~ .
##
## ── Model ───────────────────────────────────────────────────────────────────────
## Random Forest Model Specification (classification)
##
## Main Arguments:
## mtry = tune()
## trees = 500
## min_n = tune()
##
## Computational engine: ranger
Tune Random Forest Model
rf_rs_tune <- tune_grid(
object = rf_wf,
resamples = dat2_cv,
grid = 25,
control = control_resamples(save_pred = TRUE)
)
rf_rs_tune
## # Tuning results
## # 5-fold cross-validation using stratification
## # A tibble: 5 × 5
## splits id .metrics .notes .predictions
## <list> <chr> <list> <list> <list>
## 1 <split [41472/10370]> Fold1 <tibble [50 × 6]> <tibble [0 × 3]> <tibble>
## 2 <split [41474/10368]> Fold2 <tibble [50 × 6]> <tibble [0 × 3]> <tibble>
## 3 <split [41474/10368]> Fold3 <tibble [50 × 6]> <tibble [0 × 3]> <tibble>
## 4 <split [41474/10368]> Fold4 <tibble [50 × 6]> <tibble [0 × 3]> <tibble>
## 5 <split [41474/10368]> Fold5 <tibble [50 × 6]> <tibble [0 × 3]> <tibble>
# Assess performance of random forest model
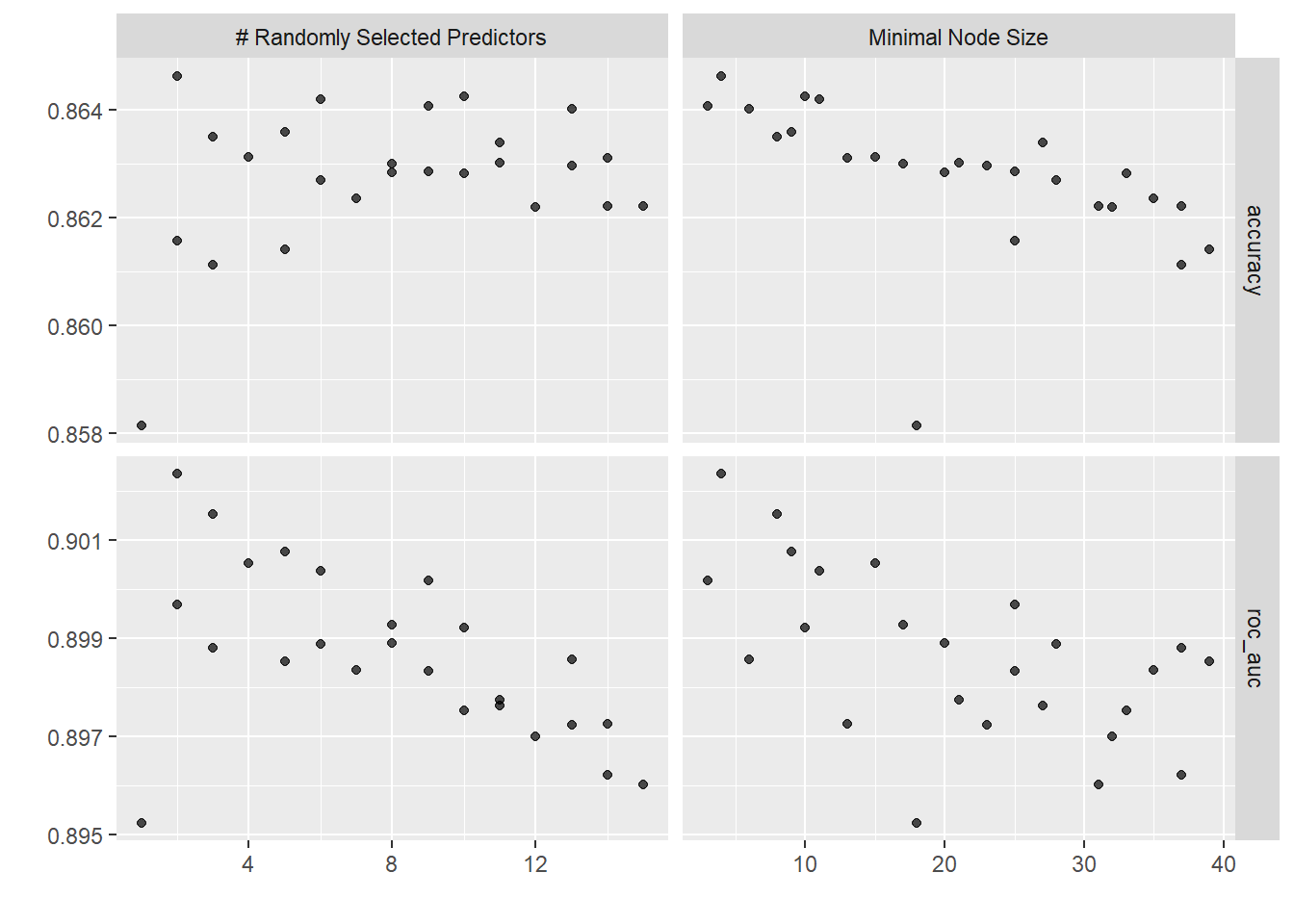
rf_rs_tune %>%
collect_metrics()
## # A tibble: 50 × 8
## mtry min_n .metric .estimator mean n std_err .config
## <int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 10 10 accuracy binary 0.864 5 0.000943 Preprocessor1_Model01
## 2 10 10 roc_auc binary 0.899 5 0.00160 Preprocessor1_Model01
## 3 11 27 accuracy binary 0.863 5 0.000626 Preprocessor1_Model02
## 4 11 27 roc_auc binary 0.898 5 0.00155 Preprocessor1_Model02
## 5 12 32 accuracy binary 0.862 5 0.000825 Preprocessor1_Model03
## 6 12 32 roc_auc binary 0.897 5 0.00162 Preprocessor1_Model03
## 7 13 6 accuracy binary 0.864 5 0.00102 Preprocessor1_Model04
## 8 13 6 roc_auc binary 0.899 5 0.00168 Preprocessor1_Model04
## 9 2 25 accuracy binary 0.862 5 0.000292 Preprocessor1_Model05
## 10 2 25 roc_auc binary 0.900 5 0.00114 Preprocessor1_Model05
## # … with 40 more rows
# Plot mtry and min_n combinations
autoplot(rf_rs_tune)
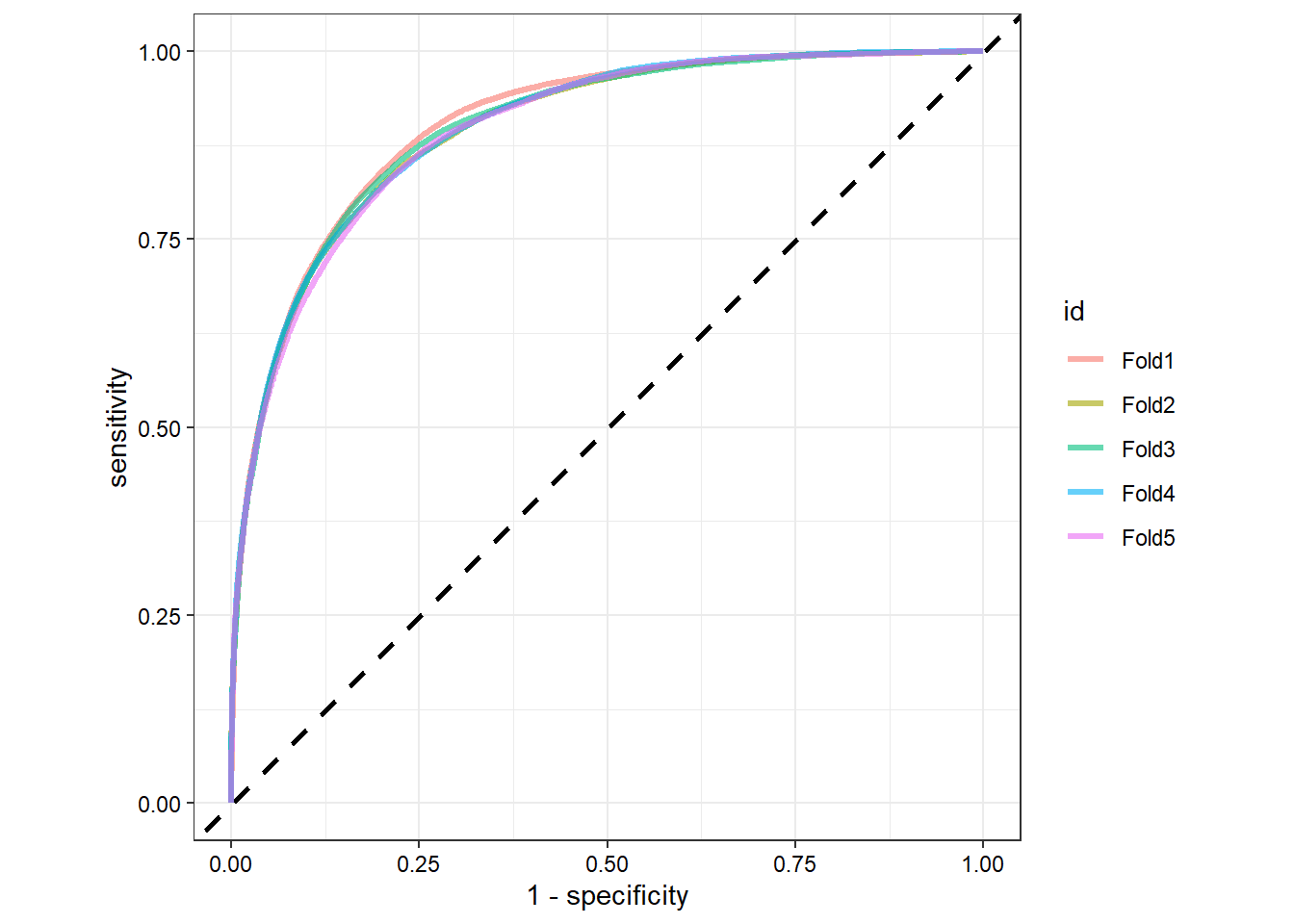
# Check sensitivity and specificity by each fold in ROC curve
rf_rs_tune %>%
collect_predictions() %>%
group_by(id) %>%
roc_curve(RainTomorrow, .pred_Yes) %>%
ggplot(aes(x = 1 - specificity, y = sensitivity, color = id)) +
geom_abline(lty = 2, color = 'black', size = 1) +
geom_path(show.legend = TRUE, alpha = 0.6, size = 1.2) +
coord_equal() +
theme_bw() +
theme(axis.text = element_text(color = 'black'))
Final Random Forest Model
# Select best model according to accuracy
best_acu <- select_best(rf_rs_tune, 'accuracy')
best_acu
## # A tibble: 1 × 3
## mtry min_n .config
## <int> <int> <chr>
## 1 2 4 Preprocessor1_Model20
# Select best model according to AUC
best_auc <- select_best(rf_rs_tune, 'roc_auc')
best_auc
## # A tibble: 1 × 3
## mtry min_n .config
## <int> <int> <chr>
## 1 2 4 Preprocessor1_Model20
# Get final model according to accuracy
rf_rs_final <- finalize_model(
rf_spec,
best_acu
)
rf_rs_final
## Random Forest Model Specification (classification)
##
## Main Arguments:
## mtry = 2
## trees = 500
## min_n = 4
##
## Computational engine: ranger
# The last_fit fits model on training data and evaluates model on test data
rf_final <- dat2_wf %>%
add_model(spec = rf_rs_final) %>%
last_fit(dat2_split)
# Check accuracy on test data
rf_final %>%
collect_metrics()
## # A tibble: 2 × 4
## .metric .estimator .estimate .config
## <chr> <chr> <dbl> <chr>
## 1 accuracy binary 0.866 Preprocessor1_Model1
## 2 roc_auc binary 0.903 Preprocessor1_Model1
# Confusion matrix on test data
rf_final %>%
collect_predictions() %>%
conf_mat(RainTomorrow, .pred_class)
## Truth
## Prediction Yes No
## Yes 2112 616
## No 1694 12860
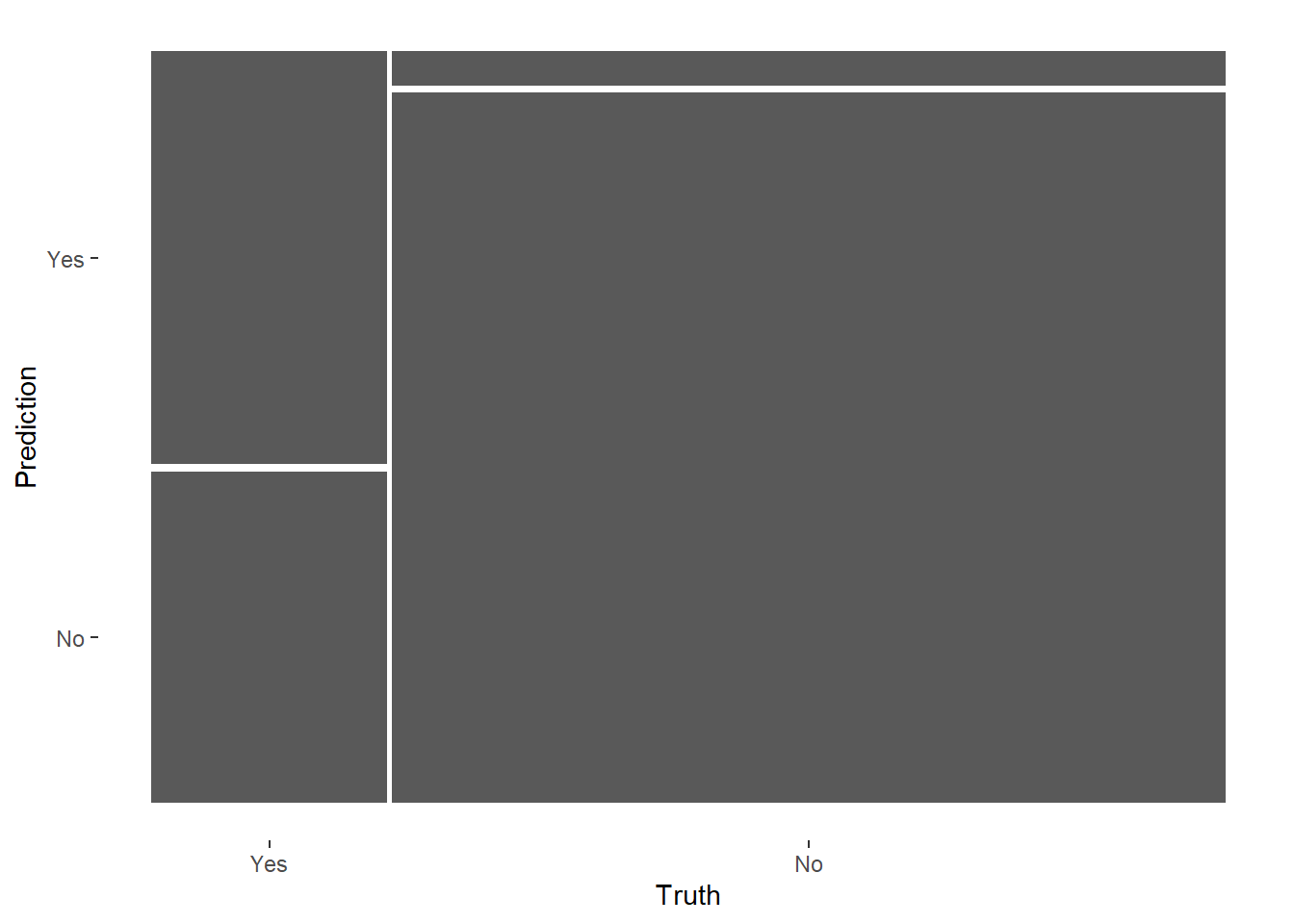
rf_final %>%
collect_predictions() %>%
conf_mat(RainTomorrow, .pred_class) %>%
autoplot(type = 'mosaic')
# Check sensitivity and specificity
rf_final %>%
collect_predictions() %>%
sensitivity(RainTomorrow, .pred_class)
## # A tibble: 1 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 sensitivity binary 0.555
rf_final %>%
collect_predictions() %>%
specificity(RainTomorrow, .pred_class)
## # A tibble: 1 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 specificity binary 0.954
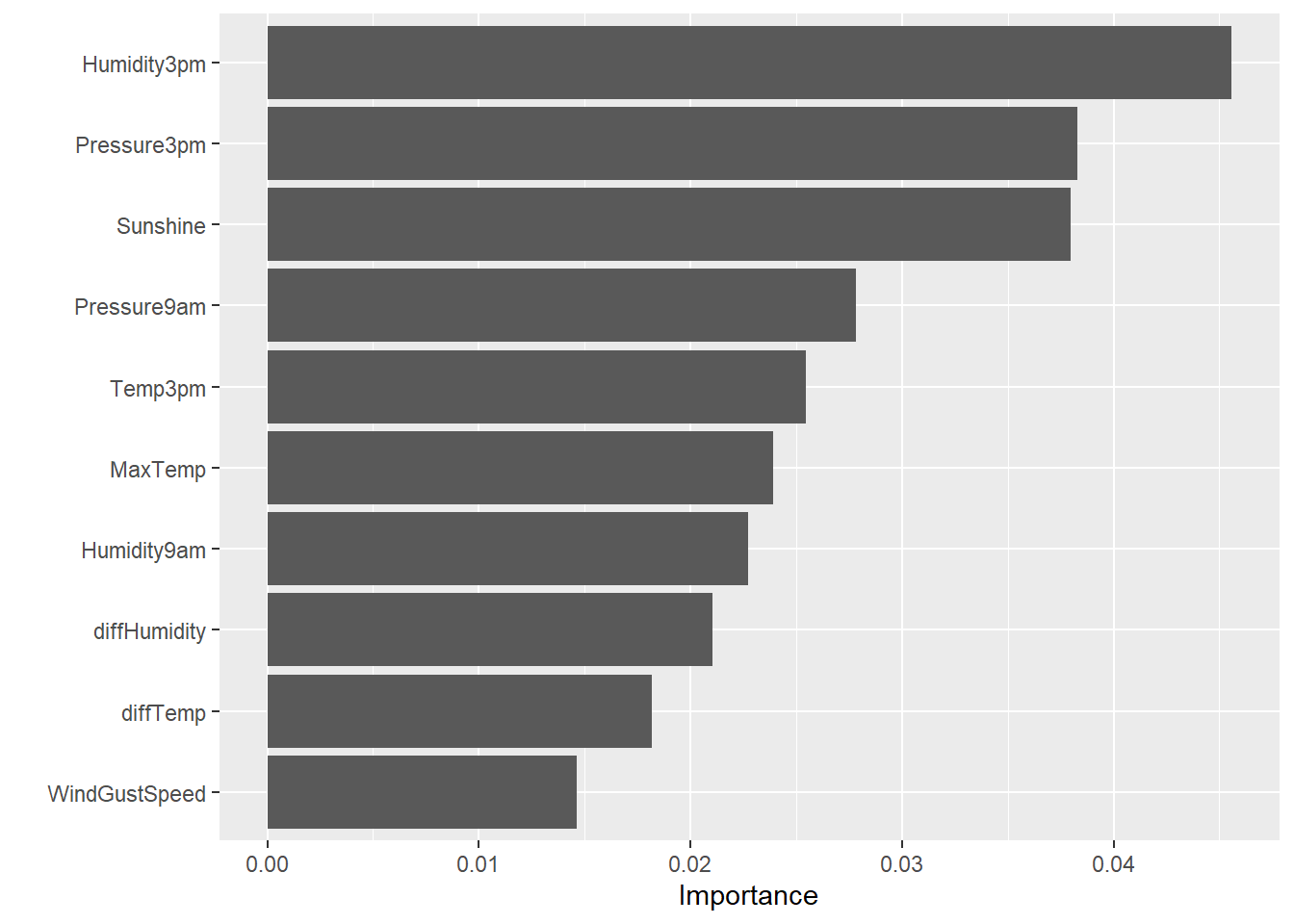
# Variable importance using vip library
rf_rs_final %>%
set_engine('ranger', importance = 'permutation') %>%
fit(RainTomorrow ~ . , data = dat2) %>%
vip(num_features = 10)
Neural Network Model
Let’s build a neural network classifier using the nnet engine. We’ll use a grid search to tune the model.
Add Neural Network Model to Workflow
# Add the neural network model to the workflow
mlp_wf <- dat2_wf %>%
add_model(mlp_spec)
mlp_wf
## ══ Workflow ════════════════════════════════════════════════════════════════════
## Preprocessor: Formula
## Model: mlp()
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## RainTomorrow ~ .
##
## ── Model ───────────────────────────────────────────────────────────────────────
## Single Layer Neural Network Model Specification (classification)
##
## Main Arguments:
## hidden_units = tune()
## penalty = tune()
## epochs = tune()
##
## Engine-Specific Arguments:
## MaxNWts = 84581
## trace = 0
##
## Computational engine: nnet
# The extract_parameter_set_dials() function can extract the set of arguments
# with unknown values and sets their dials objects
mlp_param <- extract_parameter_set_dials(mlp_spec)
mlp_param %>% extract_parameter_dials('hidden_units')
## # Hidden Units (quantitative)
## Range: [1, 10]
mlp_param %>% extract_parameter_dials('penalty')
## Amount of Regularization (quantitative)
## Transformer: log-10 [1e-100, Inf]
## Range (transformed scale): [-10, 0]
mlp_param %>% extract_parameter_dials('epochs')
## # Epochs (quantitative)
## Range: [10, 1000]
# Change number of epochs to have smaller range (50 to 200 epochs)
mlp_param <-
mlp_wf %>%
extract_parameter_set_dials() %>%
update(
epochs = epochs(c(50, 200))
)
Tune Neural Network Model
# Tune using a regular grid with three levels across the resamples
set.seed(123)
mlp_reg_tune <-
mlp_wf %>%
tune_grid(
dat2_cv,
grid = mlp_param %>% grid_regular(levels = 3),
metrics = metric_set(roc_auc)
)
mlp_reg_tune
## # Tuning results
## # 5-fold cross-validation using stratification
## # A tibble: 5 × 4
## splits id .metrics .notes
## <list> <chr> <list> <list>
## 1 <split [41472/10370]> Fold1 <tibble [27 × 7]> <tibble [0 × 3]>
## 2 <split [41474/10368]> Fold2 <tibble [27 × 7]> <tibble [0 × 3]>
## 3 <split [41474/10368]> Fold3 <tibble [27 × 7]> <tibble [0 × 3]>
## 4 <split [41474/10368]> Fold4 <tibble [27 × 7]> <tibble [0 × 3]>
## 5 <split [41474/10368]> Fold5 <tibble [27 × 7]> <tibble [0 × 3]>
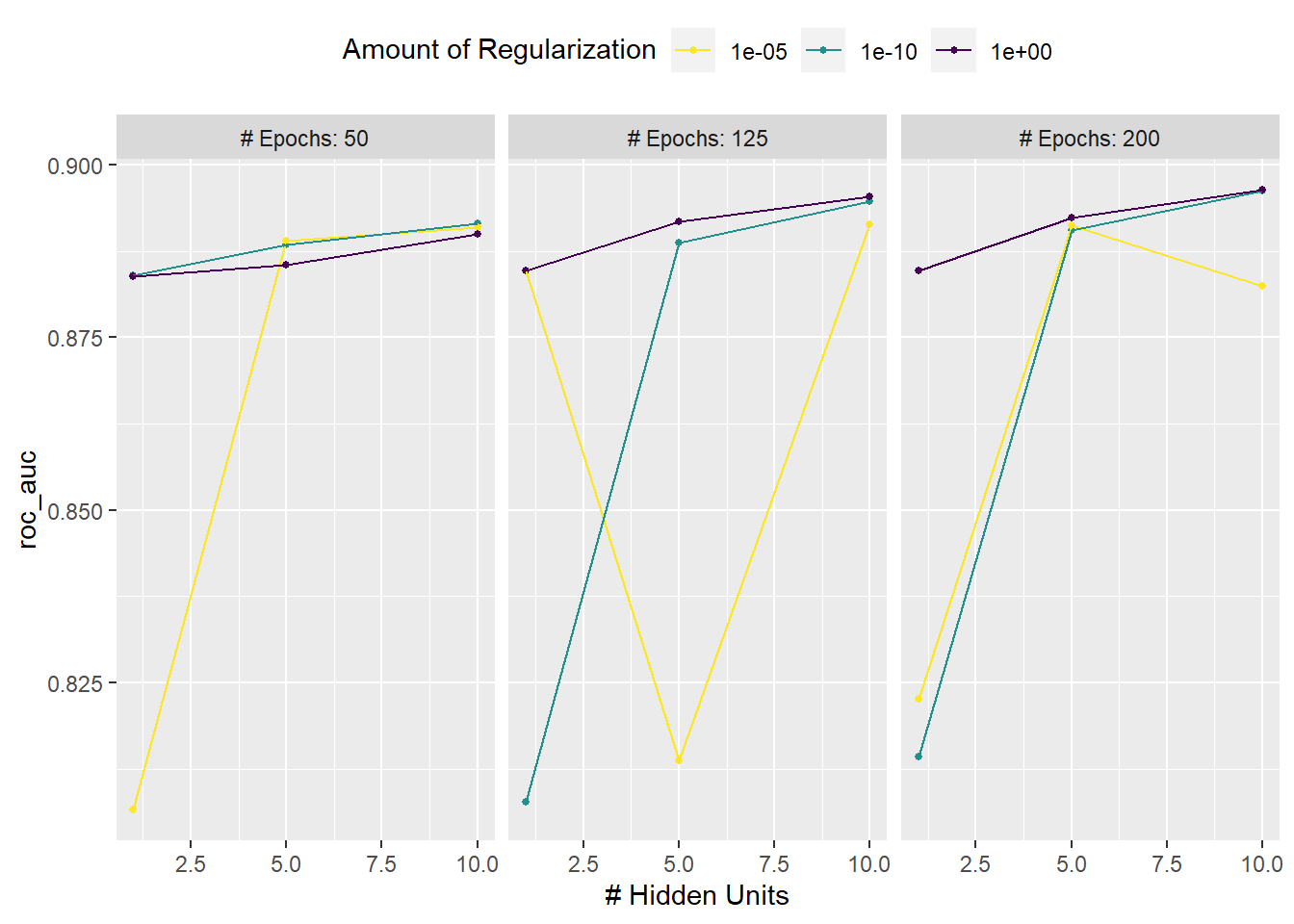
# View performance profiles across tuning parameters
autoplot(mlp_reg_tune) +
scale_color_viridis_d(direction = -1) +
theme(legend.position = 'top')
# Show best parameter configurations
show_best(mlp_reg_tune) %>% select(-.estimator)
## # A tibble: 5 × 8
## hidden_units penalty epochs .metric mean n std_err .config
## <int> <dbl> <int> <chr> <dbl> <int> <dbl> <chr>
## 1 10 1 200 roc_auc 0.896 5 0.000676 Preprocessor1_M…
## 2 10 0.0000000001 200 roc_auc 0.896 5 0.00109 Preprocessor1_M…
## 3 10 1 125 roc_auc 0.895 5 0.000984 Preprocessor1_M…
## 4 10 0.0000000001 125 roc_auc 0.895 5 0.00116 Preprocessor1_M…
## 5 5 1 200 roc_auc 0.892 5 0.000800 Preprocessor1_M…
Final Neural Network Model
# Choose parameters with numerically best result
select_best(mlp_reg_tune, metric = 'roc_auc')
## # A tibble: 1 × 4
## hidden_units penalty epochs .config
## <int> <dbl> <int> <chr>
## 1 10 1 200 Preprocessor1_Model27
# Create tibble with these values
mlp_param <-
tibble(
num_comp = 0,
epochs = 200,
hidden_units = 10,
penalty = 1
)
# Use finalize function to splice values back into workflow
final_mlp_wf <-
mlp_wf %>%
finalize_workflow(mlp_param)
final_mlp_wf
## ══ Workflow ════════════════════════════════════════════════════════════════════
## Preprocessor: Formula
## Model: mlp()
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## RainTomorrow ~ .
##
## ── Model ───────────────────────────────────────────────────────────────────────
## Single Layer Neural Network Model Specification (classification)
##
## Main Arguments:
## hidden_units = 10
## penalty = 1
## epochs = 200
##
## Engine-Specific Arguments:
## MaxNWts = 84581
## trace = 0
##
## Computational engine: nnet
# Fit model on entire training data set
mlp_final <-
final_mlp_wf %>%
fit(dat2_train)
mlp_final
## ══ Workflow [trained] ══════════════════════════════════════════════════════════
## Preprocessor: Formula
## Model: mlp()
##
## ── Preprocessor ────────────────────────────────────────────────────────────────
## RainTomorrow ~ .
##
## ── Model ───────────────────────────────────────────────────────────────────────
## a 25-10-1 network with 271 weights
## inputs: RainTodayYes Zone MonthFeb MonthMar MonthApr MonthMay MonthJun MonthJul MonthAug MonthSep MonthOct MonthNov MonthDec Sunshine diffTemp diffPressure diffHumidity WindGustSpeed MaxTemp Temp3pm Rainfall Humidity9am Humidity3pm Pressure9am Pressure3pm
## output(s): ..y
## options were - entropy fitting decay=1
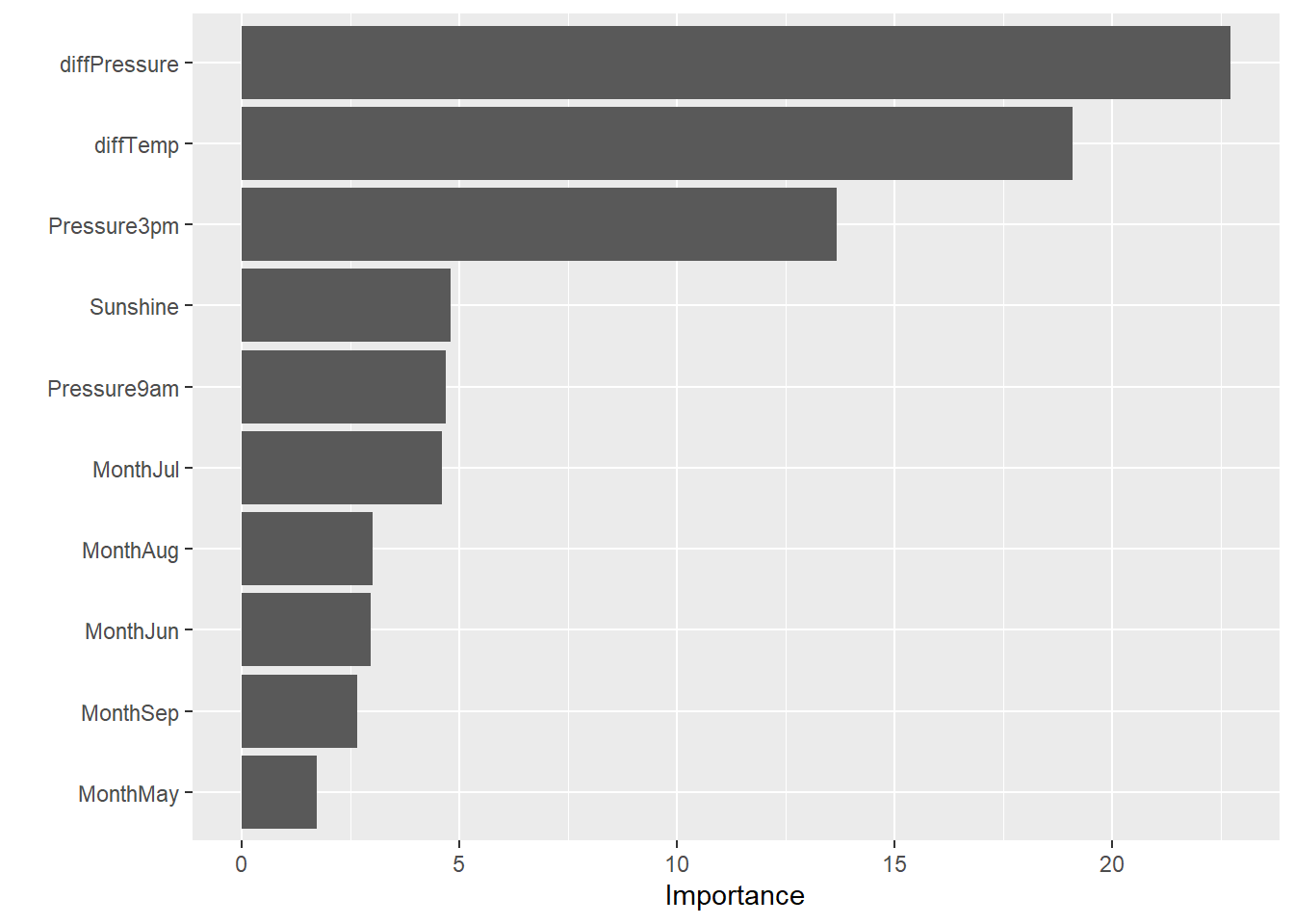
# Variable importance
mlp_final %>%
fit(data = dat2_train) %>%
extract_fit_parsnip() %>%
vip(num_features = 10)
# Check accuracy on test data set
mlp_final %>%
augment(dat2_test, type.predict = 'response') %>%
accuracy(RainTomorrow, .pred_class)
## # A tibble: 1 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 accuracy binary 0.859
# Confusion matrix on test data
mlp_final %>%
augment(dat2_test, type.predict = 'response') %>%
conf_mat(RainTomorrow, .pred_class)
## Truth
## Prediction Yes No
## Yes 2141 775
## No 1665 12701
mlp_final %>%
augment(dat2_test, type.predict = 'response') %>%
conf_mat(RainTomorrow, .pred_class) %>%
autoplot(type = 'mosaic')
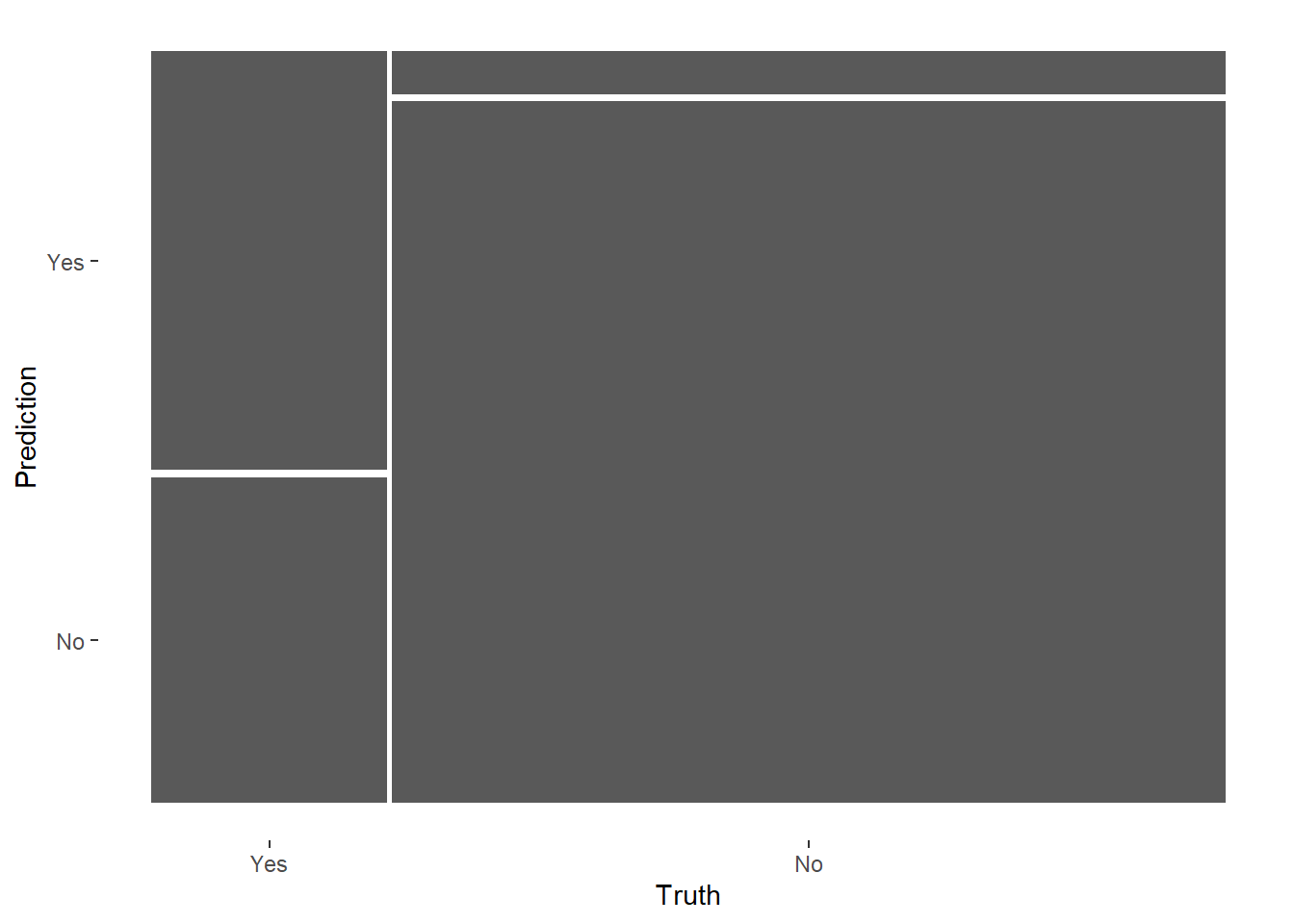
# Check sensitivity and specificity
mlp_final %>%
augment(dat2_test, type.predict = 'response') %>%
sensitivity(RainTomorrow, .pred_class)
## # A tibble: 1 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 sensitivity binary 0.563
mlp_final %>%
augment(dat2_test, type.predict = 'response') %>%
specificity(RainTomorrow, .pred_class)
## # A tibble: 1 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 specificity binary 0.942
Compare Models
Extract metrics from the four models and compare them.
glm_metrics <-
glm_final %>%
collect_metrics(summarise = TRUE) %>%
mutate(model = 'Logistic Regression')
xgb_metrics <-
xgb_final %>%
collect_metrics(summarise = TRUE) %>%
mutate(model = 'XGBoost')
rf_metrics <-
rf_final %>%
collect_metrics(summarise = TRUE) %>%
mutate(model = 'Random Forest')
mlp_metrics <-
mlp_final %>%
augment(dat2_test, type.predict = 'response') %>%
accuracy(RainTomorrow, .pred_class) %>%
bind_rows(
mlp_final %>%
augment(dat2_test, type.predict = 'prob') %>%
roc_auc(dat2_test$RainTomorrow, .pred_Yes)
) %>%
mutate(model = 'Neural Net')
# Create dataframe with all models
model_compare <- bind_rows(
glm_metrics,
rf_metrics,
xgb_metrics,
mlp_metrics
) %>%
dplyr::select(model, .metric, .estimate) %>%
pivot_wider(names_from = .metric, values_from = .estimate)
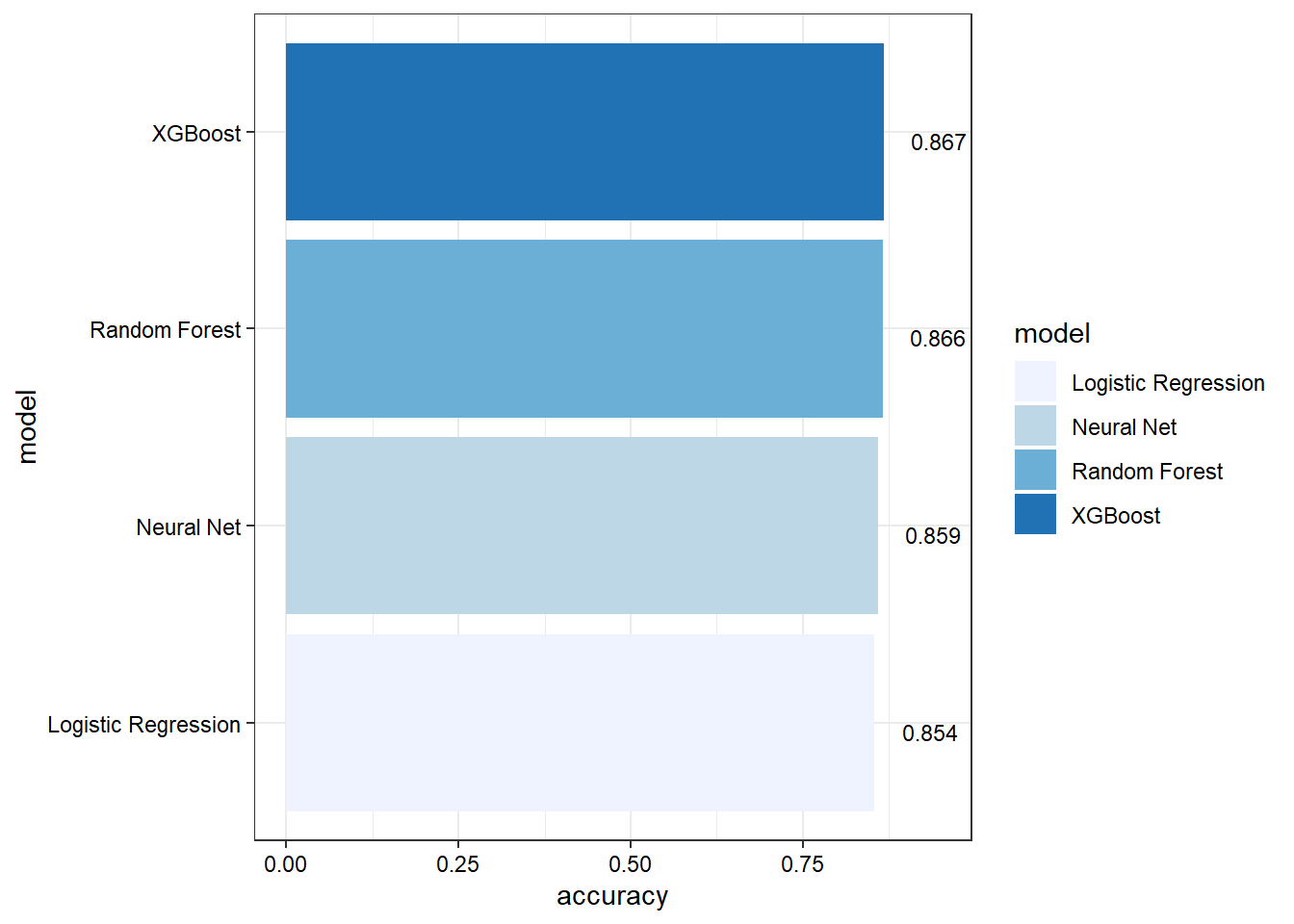
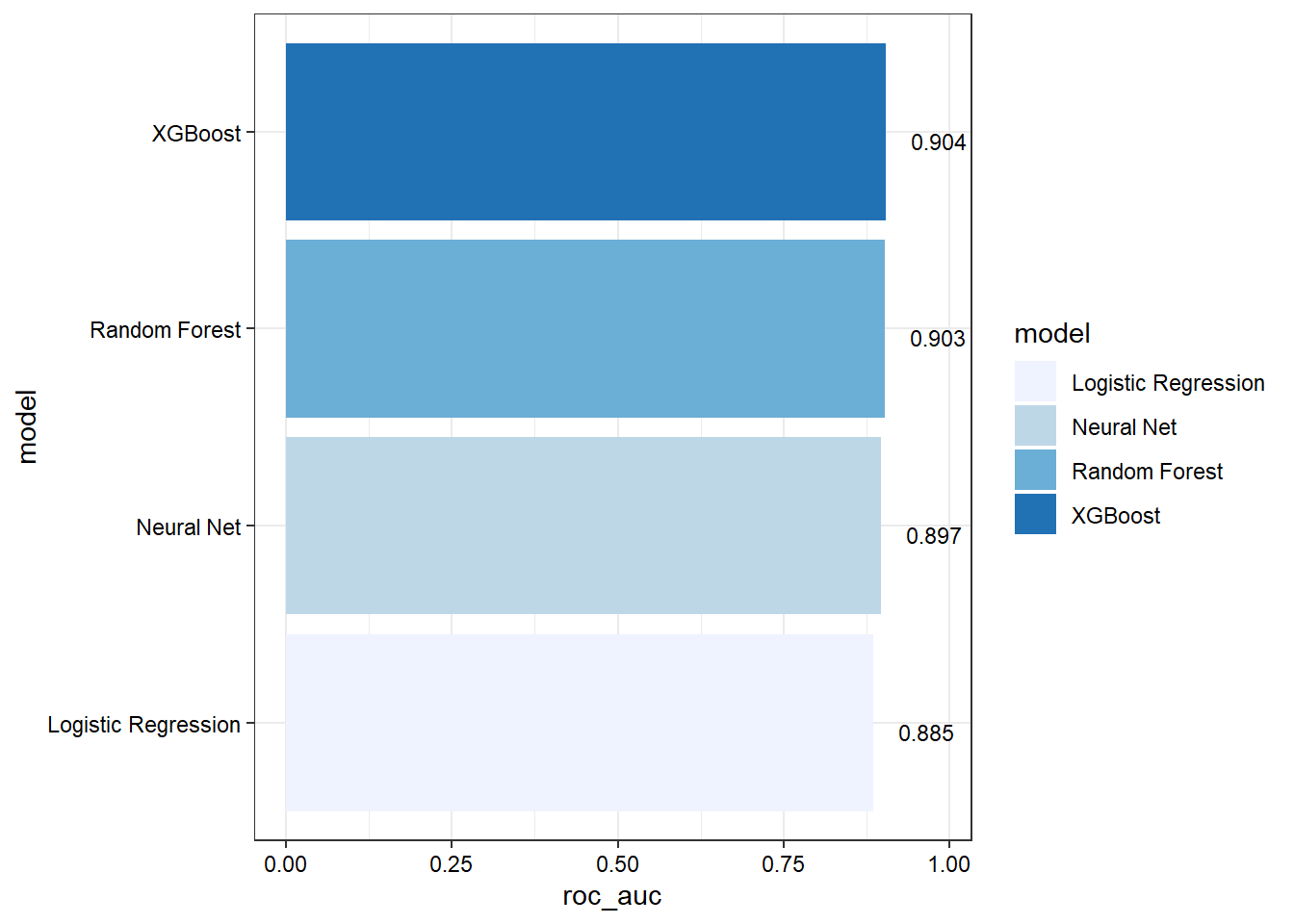
model_compare
## # A tibble: 4 × 3
## model accuracy roc_auc
## <chr> <dbl> <dbl>
## 1 Logistic Regression 0.854 0.885
## 2 Random Forest 0.866 0.903
## 3 XGBoost 0.867 0.904
## 4 Neural Net 0.859 0.897
Show accuracy for each model.
model_compare %>%
arrange(accuracy) %>%
mutate(model = forcats::fct_reorder(model, accuracy)) %>%
ggplot(aes(x = model, y = accuracy, fill = model)) +
geom_col() +
coord_flip() +
scale_fill_brewer(palette = 'Blues') +
geom_text(aes(label = round(accuracy, 3), y = accuracy + 0.08),
size = 3, vjust = 1) +
theme_bw() +
theme(axis.text = element_text(color = 'black'))
Show area under the ROC curve for each model.
model_compare %>%
arrange(roc_auc) %>%
mutate(model = forcats::fct_reorder(model, roc_auc)) %>%
ggplot(aes(x = model, y = roc_auc, fill = model)) +
geom_col() +
coord_flip() +
scale_fill_brewer(palette = 'Blues') +
geom_text(aes(label = round(roc_auc, 3), y = roc_auc + 0.08),
size = 3, vjust = 1) +
theme_bw() +
theme(axis.text = element_text(color = 'black'))
Model Findings
Based on the results, the validation set and test set performance statistics are similar across all models. Specific findings are as follows:
- Models perform good on predicting no rain tomorrow.
- Predicting it will rain tomorrow is only about 50%.
- Random forest and XGBoost perform slightly better than logistic regression and neutral network.
- Humidity, sunshine, pressure, and wind gust speed appear to be the most important variables in predicting rain tomorrow.
Further Improvement
Possible improvements to the modeling process include:
- Impute missing values.
- Try a stacked ensemble model using a GLM, random forest, and GBM model.
- Posted on:
- August 28, 2022
- Length:
- 32 minute read, 6808 words
- See Also: